Khashayar Namdar
Anti-Money Laundering Machine Learning Pipelines; A Technical Analysis on Identifying High-risk Bank Clients with Supervised Learning
Sep 11, 2025Abstract:Anti-money laundering (AML) actions and measurements are among the priorities of financial institutions, for which machine learning (ML) has shown to have a high potential. In this paper, we propose a comprehensive and systematic approach for developing ML pipelines to identify high-risk bank clients in a dataset curated for Task 1 of the University of Toronto 2023-2024 Institute for Management and Innovation (IMI) Big Data and Artificial Intelligence Competition. The dataset included 195,789 customer IDs, and we employed a 16-step design and statistical analysis to ensure the final pipeline was robust. We also framed the data in a SQLite database, developed SQL-based feature engineering algorithms, connected our pre-trained model to the database, and made it inference-ready, and provided explainable artificial intelligence (XAI) modules to derive feature importance. Our pipeline achieved a mean area under the receiver operating characteristic curve (AUROC) of 0.961 with a standard deviation (SD) of 0.005. The proposed pipeline achieved second place in the competition.
AI-Driven MRI-based Brain Tumour Segmentation Benchmarking
Jun 25, 2025Abstract:Medical image segmentation has greatly aided medical diagnosis, with U-Net based architectures and nnU-Net providing state-of-the-art performance. There have been numerous general promptable models and medical variations introduced in recent years, but there is currently a lack of evaluation and comparison of these models across a variety of prompt qualities on a common medical dataset. This research uses Segment Anything Model (SAM), Segment Anything Model 2 (SAM 2), MedSAM, SAM-Med-3D, and nnU-Net to obtain zero-shot inference on the BraTS 2023 adult glioma and pediatrics dataset across multiple prompt qualities for both points and bounding boxes. Several of these models exhibit promising Dice scores, particularly SAM and SAM 2 achieving scores of up to 0.894 and 0.893, respectively when given extremely accurate bounding box prompts which exceeds nnU-Net's segmentation performance. However, nnU-Net remains the dominant medical image segmentation network due to the impracticality of providing highly accurate prompts to the models. The model and prompt evaluation, as well as the comparison, are extended through fine-tuning SAM, SAM 2, MedSAM, and SAM-Med-3D on the pediatrics dataset. The improvements in point prompt performance after fine-tuning are substantial and show promise for future investigation, but are unable to achieve better segmentation than bounding boxes or nnU-Net.
Red Teaming Large Language Models for Healthcare
May 01, 2025Abstract:We present the design process and findings of the pre-conference workshop at the Machine Learning for Healthcare Conference (2024) entitled Red Teaming Large Language Models for Healthcare, which took place on August 15, 2024. Conference participants, comprising a mix of computational and clinical expertise, attempted to discover vulnerabilities -- realistic clinical prompts for which a large language model (LLM) outputs a response that could cause clinical harm. Red-teaming with clinicians enables the identification of LLM vulnerabilities that may not be recognised by LLM developers lacking clinical expertise. We report the vulnerabilities found, categorise them, and present the results of a replication study assessing the vulnerabilities across all LLMs provided.
Opportunities for Persian Digital Humanities Research with Artificial Intelligence Language Models; Case Study: Forough Farrokhzad
May 10, 2024



Abstract:This study explores the integration of advanced Natural Language Processing (NLP) and Artificial Intelligence (AI) techniques to analyze and interpret Persian literature, focusing on the poetry of Forough Farrokhzad. Utilizing computational methods, we aim to unveil thematic, stylistic, and linguistic patterns in Persian poetry. Specifically, the study employs AI models including transformer-based language models for clustering of the poems in an unsupervised framework. This research underscores the potential of AI in enhancing our understanding of Persian literary heritage, with Forough Farrokhzad's work providing a comprehensive case study. This approach not only contributes to the field of Persian Digital Humanities but also sets a precedent for future research in Persian literary studies using computational techniques.
Improving Pediatric Low-Grade Neuroepithelial Tumors Molecular Subtype Identification Using a Novel AUROC Loss Function for Convolutional Neural Networks
Feb 05, 2024

Abstract:Pediatric Low-Grade Neuroepithelial Tumors (PLGNT) are the most common pediatric cancer type, accounting for 40% of brain tumors in children, and identifying PLGNT molecular subtype is crucial for treatment planning. However, the gold standard to determine the PLGNT subtype is biopsy, which can be impractical or dangerous for patients. This research improves the performance of Convolutional Neural Networks (CNNs) in classifying PLGNT subtypes through MRI scans by introducing a loss function that specifically improves the model's Area Under the Receiver Operating Characteristic (ROC) Curve (AUROC), offering a non-invasive diagnostic alternative. In this study, a retrospective dataset of 339 children with PLGNT (143 BRAF fusion, 71 with BRAF V600E mutation, and 125 non-BRAF) was curated. We employed a CNN model with Monte Carlo random data splitting. The baseline model was trained using binary cross entropy (BCE), and achieved an AUROC of 86.11% for differentiating BRAF fusion and BRAF V600E mutations, which was improved to 87.71% using our proposed AUROC loss function (p-value 0.045). With multiclass classification, the AUROC improved from 74.42% to 76. 59% (p-value 0.0016).
Automating Cobb Angle Measurement for Adolescent Idiopathic Scoliosis using Instance Segmentation
Nov 25, 2022Abstract:Scoliosis is a three-dimensional deformity of the spine, most often diagnosed in childhood. It affects 2-3% of the population, which is approximately seven million people in North America. Currently, the reference standard for assessing scoliosis is based on the manual assignment of Cobb angles at the site of the curvature center. This manual process is time consuming and unreliable as it is affected by inter- and intra-observer variance. To overcome these inaccuracies, machine learning (ML) methods can be used to automate the Cobb angle measurement process. This paper proposes to address the Cobb angle measurement task using YOLACT, an instance segmentation model. The proposed method first segments the vertebrae in an X-Ray image using YOLACT, then it tracks the important landmarks using the minimum bounding box approach. Lastly, the extracted landmarks are used to calculate the corresponding Cobb angles. The model achieved a Symmetric Mean Absolute Percentage Error (SMAPE) score of 10.76%, demonstrating the reliability of this process in both vertebra localization and Cobb angle measurement.
A Comprehensive Study of Radiomics-based Machine Learning for Fibrosis Detection
Nov 25, 2022Abstract:Objectives: Early detection of liver fibrosis can help cure the disease or prevent disease progression. We perform a comprehensive study of machine learning-based fibrosis detection in CT images using radiomic features to develop a non-invasive approach to fibrosis detection. Methods: Two sets of radiomic features were extracted from spherical ROIs in CT images of 182 patients who underwent simultaneous liver biopsy and CT examinations, one set corresponding to biopsy locations and another distant from biopsy locations. Combinations of contrast, normalization, machine learning model, feature selection method, bin width, and kernel radius were investigated, each of which were trained and evaluated 100 times with randomized development and test cohorts. The best settings were evaluated based on their mean test AUC and the best features were determined based on their frequency among the best settings. Results: Logistic regression models with NC images normalized using Gamma correction with $\gamma = 1.5$ performed best for fibrosis detection. Boruta was the best for radiomic feature selection method. Training a model using these optimal settings and features consisting of first order energy, first order kurtosis, and first order skewness, resulted in a model that achieved mean test AUCs of 0.7549 and 0.7166 on biopsy-based and non-biopsy ROIs respectively, outperforming a baseline and best models found during the initial study. Conclusions: Logistic regression models trained on radiomic features from NC images normalized using Gamma correction with $\gamma = 1.5$ that underwent Boruta feature selection are effective for liver fibrosis detection. Energy, kurtosis, and skewness are particularly effective features for fibrosis detection.
A novel GAN-based paradigm for weakly supervised brain tumor segmentation of MR images
Nov 10, 2022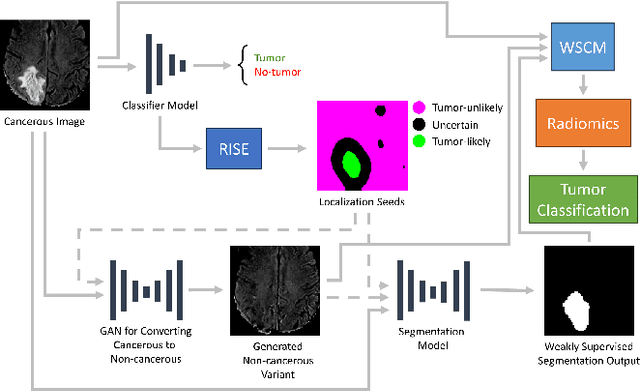
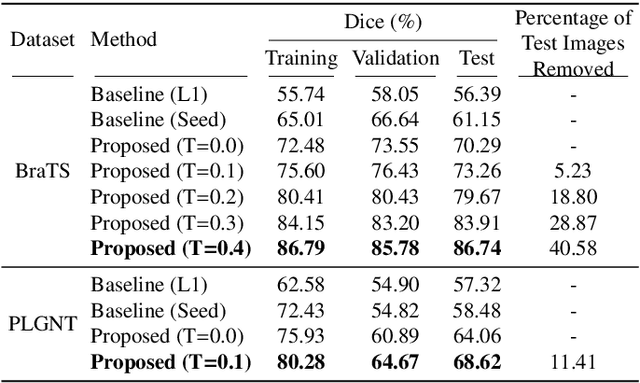
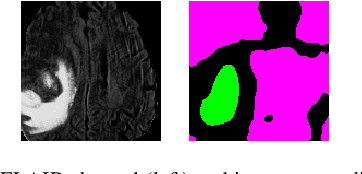
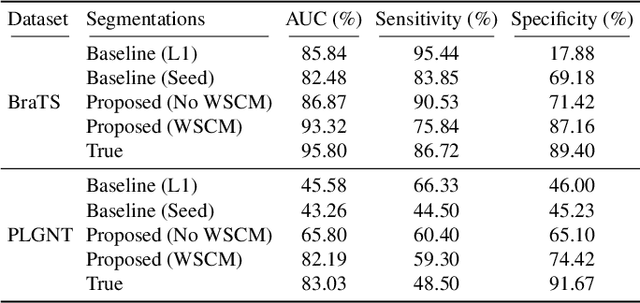
Abstract:Segmentation of regions of interest (ROIs) for identifying abnormalities is a leading problem in medical imaging. Using Machine Learning (ML) for this problem generally requires manually annotated ground-truth segmentations, demanding extensive time and resources from radiologists. This work presents a novel weakly supervised approach that utilizes binary image-level labels, which are much simpler to acquire, to effectively segment anomalies in medical Magnetic Resonance (MR) images without ground truth annotations. We train a binary classifier using these labels and use it to derive seeds indicating regions likely and unlikely to contain tumors. These seeds are used to train a generative adversarial network (GAN) that converts cancerous images to healthy variants, which are then used in conjunction with the seeds to train a ML model that generates effective segmentations. This method produces segmentations that achieve Dice coefficients of 0.7903, 0.7868, and 0.7712 on the MICCAI Brain Tumor Segmentation (BraTS) 2020 dataset for the training, validation, and test cohorts respectively. We also propose a weakly supervised means of filtering the segmentations, removing a small subset of poorer segmentations to acquire a large subset of high quality segmentations. The proposed filtering further improves the Dice coefficients to up to 0.8374, 0.8232, and 0.8136 for training, validation, and test, respectively.
Tumor-location-guided CNNs for Pediatric Low-grade Glioma Molecular Biomarker Classification Using MRI
Oct 13, 2022


Abstract:Pediatric low-grade glioma (pLGG) is the most common type of brain cancer among children, and the identification of molecular markers for pLGG is crucial for successful treatment planning. Current standard care is biopsy, which is invasive. Thus, the non-invasive imaging-based approaches, where Machine Learning (ML) has a high potential, are impactful. Recently, we developed a tumor-location-based algorithm and demonstrated its potential to differentiate pLGG molecular subtypes. In this work, we first reevaluated the performance of the location-based algorithm on a larger pLGG dataset, which includes 214 patients and achieved an area under the receiver operating characteristic curve (AUROC) of 77.90. A Convolutional Neural Network (CNN) based algorithm increased the average AUROC to 86.11. Ultimately, we designed and implemented a tumor-location-guided CNN algorithm and achieved average AUROC of 88.64. Using a repeated experiment approach with 100 runs, we ensured the results were reproducible and the improvement was statistically significant.
Superpixel Generation and Clustering for Weakly Supervised Brain Tumor Segmentation in MR Images
Sep 20, 2022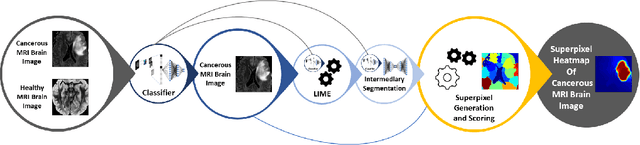
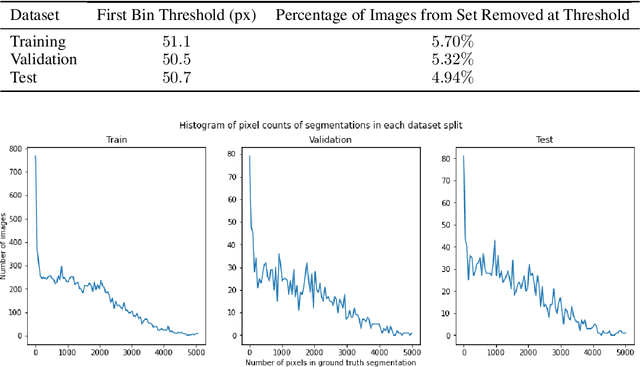
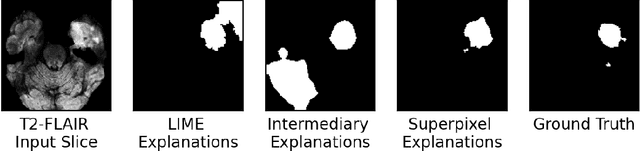
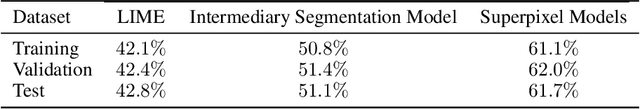
Abstract:Training Machine Learning (ML) models to segment tumors and other anomalies in medical images is an increasingly popular area of research but generally requires manually annotated ground truth segmentations which necessitates significant time and resources to create. This work proposes a pipeline of ML models that utilize binary classification labels, which can be easily acquired, to segment ROIs without requiring ground truth annotations. We used 2D slices of Magnetic Resonance Imaging (MRI) brain scans from the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 dataset and labels indicating the presence of high-grade glioma (HGG) tumors to train the pipeline. Our pipeline also introduces a novel variation of deep learning-based superpixel generation, which enables training guided by clustered superpixels and simultaneously trains a superpixel clustering model. On our test set, our pipeline's segmentations achieved a Dice coefficient of 61.7%, which is a substantial improvement over the 42.8% Dice coefficient acquired when the popular Local Interpretable Model-Agnostic Explanations (LIME) method was used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge