Han Liu
Johns Hopkins University
VAMOS-OCTA: Vessel-Aware Multi-Axis Orthogonal Supervision for Inpainting Motion-Corrupted OCT Angiography Volumes
Feb 01, 2026Abstract:Handheld Optical Coherence Tomography Angiography (OCTA) enables noninvasive retinal imaging in uncooperative or pediatric subjects, but is highly susceptible to motion artifacts that severely degrade volumetric image quality. Sudden motion during 3D acquisition can lead to unsampled retinal regions across entire B-scans (cross-sectional slices), resulting in blank bands in en face projections. We propose VAMOS-OCTA, a deep learning framework for inpainting motion-corrupted B-scans using vessel-aware multi-axis supervision. We employ a 2.5D U-Net architecture that takes a stack of neighboring B-scans as input to reconstruct a corrupted center B-scan, guided by a novel Vessel-Aware Multi-Axis Orthogonal Supervision (VAMOS) loss. This loss combines vessel-weighted intensity reconstruction with axial and lateral projection consistency, encouraging vascular continuity in native B-scans and across orthogonal planes. Unlike prior work that focuses primarily on restoring the en face MIP, VAMOS-OCTA jointly enhances both cross-sectional B-scan sharpness and volumetric projection accuracy, even under severe motion corruptions. We trained our model on both synthetic and real-world corrupted volumes and evaluated its performance using both perceptual quality and pixel-wise accuracy metrics. VAMOS-OCTA consistently outperforms prior methods, producing reconstructions with sharp capillaries, restored vessel continuity, and clean en face projections. These results demonstrate that multi-axis supervision offers a powerful constraint for restoring motion-degraded 3D OCTA data. Our source code is available at https://github.com/MedICL-VU/VAMOS-OCTA.
PROGRESSLM: Towards Progress Reasoning in Vision-Language Models
Jan 21, 2026Abstract:Estimating task progress requires reasoning over long-horizon dynamics rather than recognizing static visual content. While modern Vision-Language Models (VLMs) excel at describing what is visible, it remains unclear whether they can infer how far a task has progressed from partial observations. To this end, we introduce Progress-Bench, a benchmark for systematically evaluating progress reasoning in VLMs. Beyond benchmarking, we further explore a human-inspired two-stage progress reasoning paradigm through both training-free prompting and training-based approach based on curated dataset ProgressLM-45K. Experiments on 14 VLMs show that most models are not yet ready for task progress estimation, exhibiting sensitivity to demonstration modality and viewpoint changes, as well as poor handling of unanswerable cases. While training-free prompting that enforces structured progress reasoning yields limited and model-dependent gains, the training-based ProgressLM-3B achieves consistent improvements even at a small model scale, despite being trained on a task set fully disjoint from the evaluation tasks. Further analyses reveal characteristic error patterns and clarify when and why progress reasoning succeeds or fails.
Learning Semantic-Geometric Task Graph-Representations from Human Demonstrations
Jan 16, 2026Abstract:Learning structured task representations from human demonstrations is essential for understanding long-horizon manipulation behaviors, particularly in bimanual settings where action ordering, object involvement, and interaction geometry can vary significantly. A key challenge lies in jointly capturing the discrete semantic structure of tasks and the temporal evolution of object-centric geometric relations in a form that supports reasoning over task progression. In this work, we introduce a semantic-geometric task graph-representation that encodes object identities, inter-object relations, and their temporal geometric evolution from human demonstrations. Building on this formulation, we propose a learning framework that combines a Message Passing Neural Network (MPNN) encoder with a Transformer-based decoder, decoupling scene representation learning from action-conditioned reasoning about task progression. The encoder operates solely on temporal scene graphs to learn structured representations, while the decoder conditions on action-context to predict future action sequences, associated objects, and object motions over extended time horizons. Through extensive evaluation on human demonstration datasets, we show that semantic-geometric task graph-representations are particularly beneficial for tasks with high action and object variability, where simpler sequence-based models struggle to capture task progression. Finally, we demonstrate that task graph representations can be transferred to a physical bimanual robot and used for online action selection, highlighting their potential as reusable task abstractions for downstream decision-making in manipulation systems.
UserLM-R1: Modeling Human Reasoning in User Language Models with Multi-Reward Reinforcement Learning
Jan 14, 2026Abstract:User simulators serve as the critical interactive environment for agent post-training, and an ideal user simulator generalizes across domains and proactively engages in negotiation by challenging or bargaining. However, current methods exhibit two issues. They rely on static and context-unaware profiles, necessitating extensive manual redesign for new scenarios, thus limiting generalizability. Moreover, they neglect human strategic thinking, leading to vulnerability to agent manipulation. To address these issues, we propose UserLM-R1, a novel user language model with reasoning capability. Specifically, we first construct comprehensive user profiles with both static roles and dynamic scenario-specific goals for adaptation to diverse scenarios. Then, we propose a goal-driven decision-making policy to generate high-quality rationales before producing responses, and further refine the reasoning and improve strategic capabilities with supervised fine-tuning and multi-reward reinforcement learning. Extensive experimental results demonstrate that UserLM-R1 outperforms competitive baselines, particularly on the more challenging adversarial set.
Efficient Paths and Dense Rewards: Probabilistic Flow Reasoning for Large Language Models
Jan 14, 2026Abstract:High-quality chain-of-thought has demonstrated strong potential for unlocking the reasoning capabilities of large language models. However, current paradigms typically treat the reasoning process as an indivisible sequence, lacking an intrinsic mechanism to quantify step-wise information gain. This granularity gap manifests in two limitations: inference inefficiency from redundant exploration without explicit guidance, and optimization difficulty due to sparse outcome supervision or costly external verifiers. In this work, we propose CoT-Flow, a framework that reconceptualizes discrete reasoning steps as a continuous probabilistic flow, quantifying the contribution of each step toward the ground-truth answer. Built on this formulation, CoT-Flow enables two complementary methodologies: flow-guided decoding, which employs a greedy flow-based decoding strategy to extract information-efficient reasoning paths, and flow-based reinforcement learning, which constructs a verifier-free dense reward function. Experiments on challenging benchmarks demonstrate that CoT-Flow achieves a superior balance between inference efficiency and reasoning performance.
IntraStyler: Exemplar-based Style Synthesis for Cross-modality Domain Adaptation
Jan 01, 2026Abstract:Image-level domain alignment is the de facto approach for unsupervised domain adaptation, where unpaired image translation is used to minimize the domain gap. Prior studies mainly focus on the domain shift between the source and target domains, whereas the intra-domain variability remains under-explored. To address the latter, an effective strategy is to diversify the styles of the synthetic target domain data during image translation. However, previous methods typically require intra-domain variations to be pre-specified for style synthesis, which may be impractical. In this paper, we propose an exemplar-based style synthesis method named IntraStyler, which can capture diverse intra-domain styles without any prior knowledge. Specifically, IntraStyler uses an exemplar image to guide the style synthesis such that the output style matches the exemplar style. To extract the style-only features, we introduce a style encoder to learn styles discriminatively based on contrastive learning. We evaluate the proposed method on the largest public dataset for cross-modality domain adaptation, CrossMoDA 2023. Our experiments show the efficacy of our method in controllable style synthesis and the benefits of diverse synthetic data for downstream segmentation. Code is available at https://github.com/han-liu/IntraStyler.
Learning to Feel the Future: DreamTacVLA for Contact-Rich Manipulation
Dec 29, 2025Abstract:Vision-Language-Action (VLA) models have shown remarkable generalization by mapping web-scale knowledge to robotic control, yet they remain blind to physical contact. Consequently, they struggle with contact-rich manipulation tasks that require reasoning about force, texture, and slip. While some approaches incorporate low-dimensional tactile signals, they fail to capture the high-resolution dynamics essential for such interactions. To address this limitation, we introduce DreamTacVLA, a framework that grounds VLA models in contact physics by learning to feel the future. Our model adopts a hierarchical perception scheme in which high-resolution tactile images serve as micro-vision inputs coupled with wrist-camera local vision and third-person macro vision. To reconcile these multi-scale sensory streams, we first train a unified policy with a Hierarchical Spatial Alignment (HSA) loss that aligns tactile tokens with their spatial counterparts in the wrist and third-person views. To further deepen the model's understanding of fine-grained contact dynamics, we finetune the system with a tactile world model that predicts future tactile signals. To mitigate tactile data scarcity and the wear-prone nature of tactile sensors, we construct a hybrid large-scale dataset sourced from both high-fidelity digital twin and real-world experiments. By anticipating upcoming tactile states, DreamTacVLA acquires a rich model of contact physics and conditions its actions on both real observations and imagined consequences. Across contact-rich manipulation tasks, it outperforms state-of-the-art VLA baselines, achieving up to 95% success, highlighting the importance of understanding physical contact for robust, touch-aware robotic agents.
HY-Motion 1.0: Scaling Flow Matching Models for Text-To-Motion Generation
Dec 29, 2025Abstract:We present HY-Motion 1.0, a series of state-of-the-art, large-scale, motion generation models capable of generating 3D human motions from textual descriptions. HY-Motion 1.0 represents the first successful attempt to scale up Diffusion Transformer (DiT)-based flow matching models to the billion-parameter scale within the motion generation domain, delivering instruction-following capabilities that significantly outperform current open-source benchmarks. Uniquely, we introduce a comprehensive, full-stage training paradigm -- including large-scale pretraining on over 3,000 hours of motion data, high-quality fine-tuning on 400 hours of curated data, and reinforcement learning from both human feedback and reward models -- to ensure precise alignment with the text instruction and high motion quality. This framework is supported by our meticulous data processing pipeline, which performs rigorous motion cleaning and captioning. Consequently, our model achieves the most extensive coverage, spanning over 200 motion categories across 6 major classes. We release HY-Motion 1.0 to the open-source community to foster future research and accelerate the transition of 3D human motion generation models towards commercial maturity.
E-Navi: Environmental Adaptive Navigation for UAVs on Resource Constrained Platforms
Dec 16, 2025
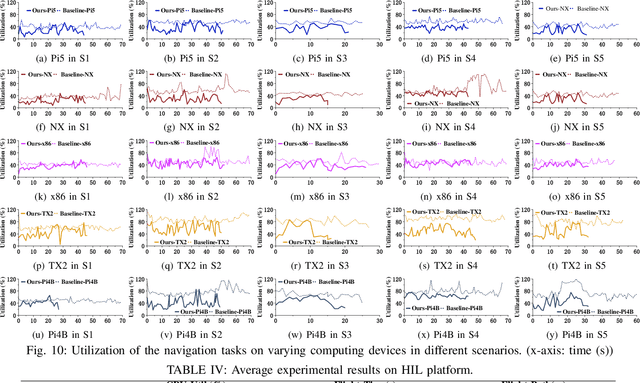
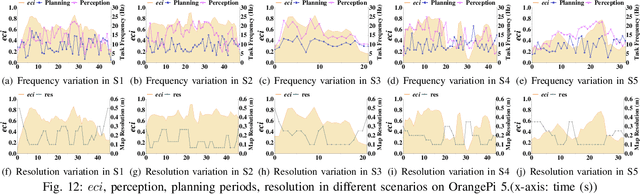
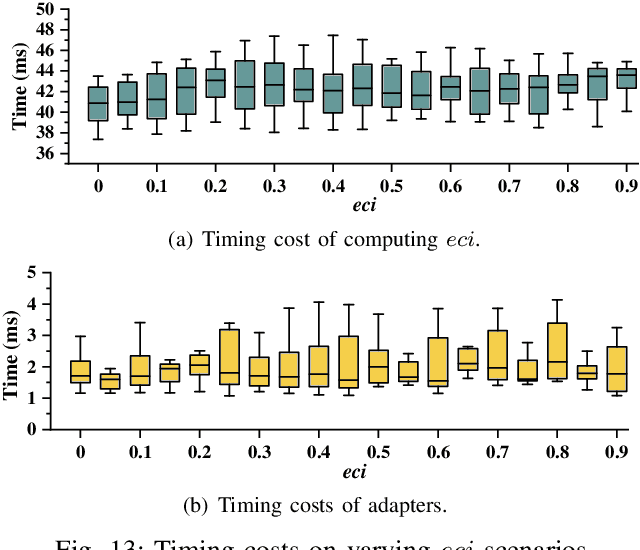
Abstract:The ability to adapt to changing environments is crucial for the autonomous navigation systems of Unmanned Aerial Vehicles (UAVs). However, existing navigation systems adopt fixed execution configurations without considering environmental dynamics based on available computing resources, e.g., with a high execution frequency and task workload. This static approach causes rigid flight strategies and excessive computations, ultimately degrading flight performance or even leading to failures in UAVs. Despite the necessity for an adaptive system, dynamically adjusting workloads remains challenging, due to difficulties in quantifying environmental complexity and modeling the relationship between environment and system configuration. Aiming at adapting to dynamic environments, this paper proposes E-Navi, an environmental-adaptive navigation system for UAVs that dynamically adjusts task executions on the CPUs in response to environmental changes based on available computational resources. Specifically, the perception-planning pipeline of UAVs navigation system is redesigned through dynamic adaptation of mapping resolution and execution frequency, driven by the quantitative environmental complexity evaluations. In addition, E-Navi supports flexible deployment across hardware platforms with varying levels of computing capability. Extensive Hardware-In-the-Loop and real-world experiments demonstrate that the proposed system significantly outperforms the baseline method across various hardware platforms, achieving up to 53.9% navigation task workload reduction, up to 63.8% flight time savings, and delivering more stable velocity control.
Revisiting 2D Foundation Models for Scalable 3D Medical Image Classification
Dec 15, 2025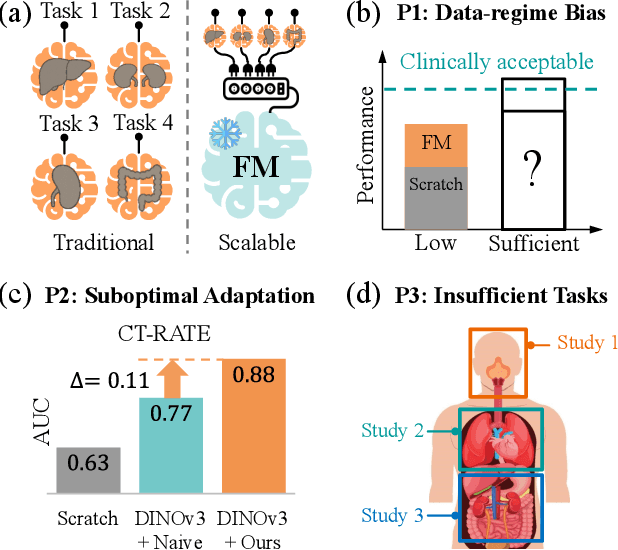
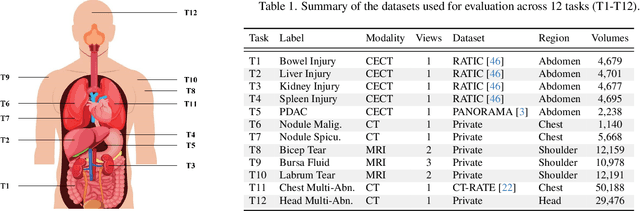
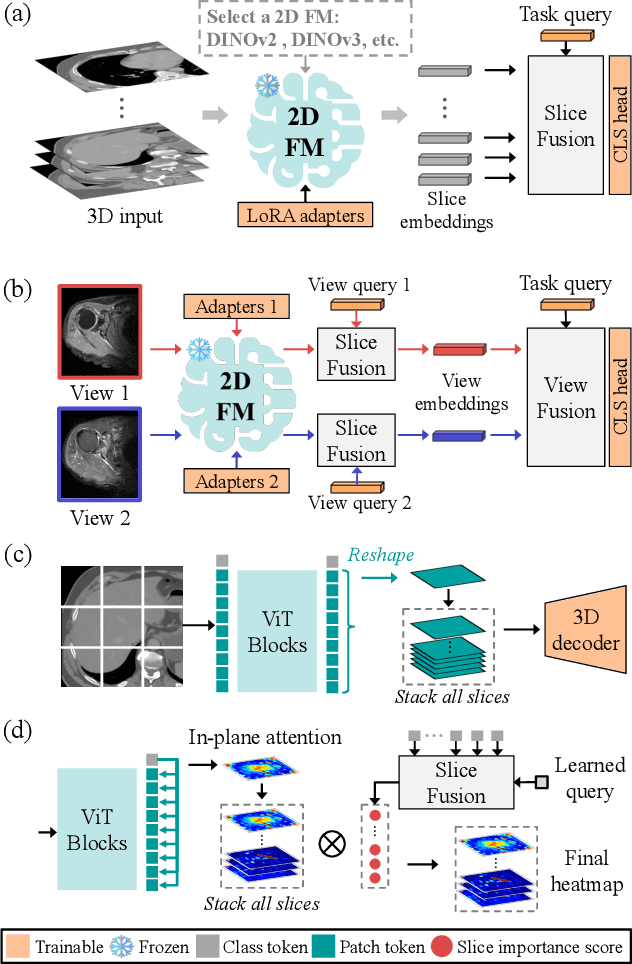
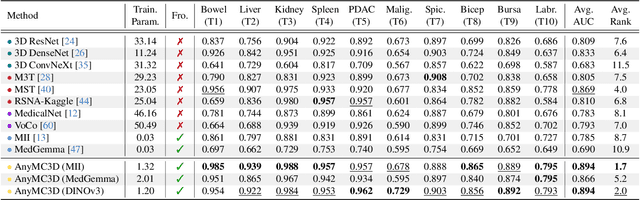
Abstract:3D medical image classification is essential for modern clinical workflows. Medical foundation models (FMs) have emerged as a promising approach for scaling to new tasks, yet current research suffers from three critical pitfalls: data-regime bias, suboptimal adaptation, and insufficient task coverage. In this paper, we address these pitfalls and introduce AnyMC3D, a scalable 3D classifier adapted from 2D FMs. Our method scales efficiently to new tasks by adding only lightweight plugins (about 1M parameters per task) on top of a single frozen backbone. This versatile framework also supports multi-view inputs, auxiliary pixel-level supervision, and interpretable heatmap generation. We establish a comprehensive benchmark of 12 tasks covering diverse pathologies, anatomies, and modalities, and systematically analyze state-of-the-art 3D classification techniques. Our analysis reveals key insights: (1) effective adaptation is essential to unlock FM potential, (2) general-purpose FMs can match medical-specific FMs if properly adapted, and (3) 2D-based methods surpass 3D architectures for 3D classification. For the first time, we demonstrate the feasibility of achieving state-of-the-art performance across diverse applications using a single scalable framework (including 1st place in the VLM3D challenge), eliminating the need for separate task-specific models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge