Gengyan Zhao
Revisiting 2D Foundation Models for Scalable 3D Medical Image Classification
Dec 15, 2025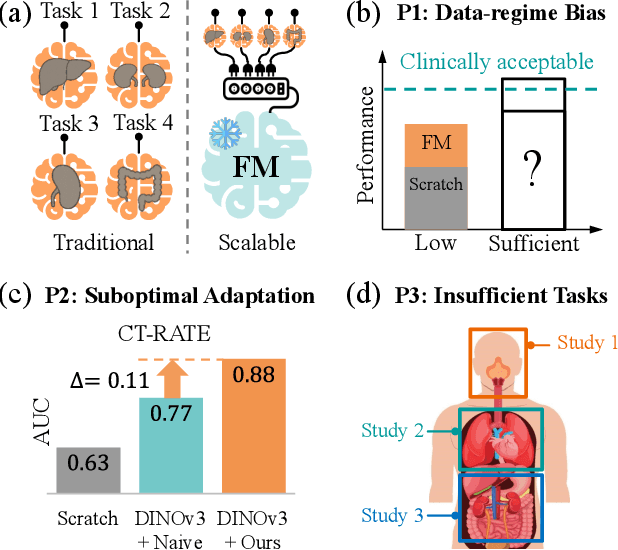
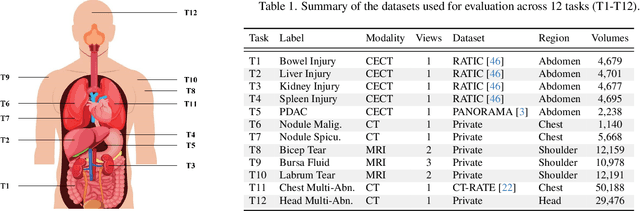
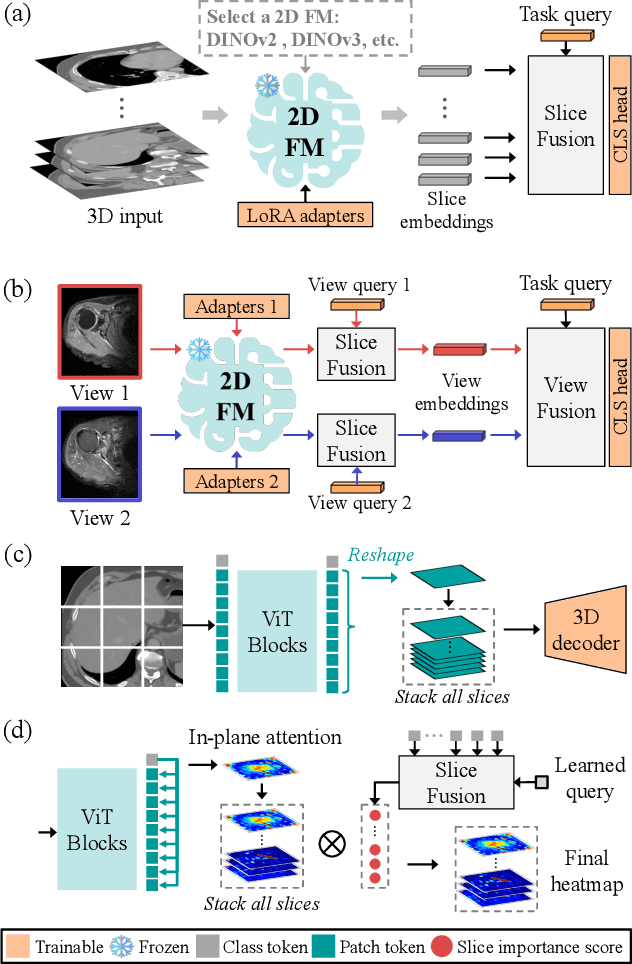
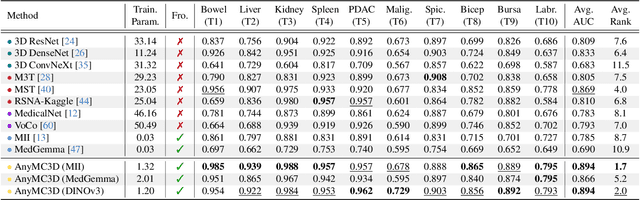
Abstract:3D medical image classification is essential for modern clinical workflows. Medical foundation models (FMs) have emerged as a promising approach for scaling to new tasks, yet current research suffers from three critical pitfalls: data-regime bias, suboptimal adaptation, and insufficient task coverage. In this paper, we address these pitfalls and introduce AnyMC3D, a scalable 3D classifier adapted from 2D FMs. Our method scales efficiently to new tasks by adding only lightweight plugins (about 1M parameters per task) on top of a single frozen backbone. This versatile framework also supports multi-view inputs, auxiliary pixel-level supervision, and interpretable heatmap generation. We establish a comprehensive benchmark of 12 tasks covering diverse pathologies, anatomies, and modalities, and systematically analyze state-of-the-art 3D classification techniques. Our analysis reveals key insights: (1) effective adaptation is essential to unlock FM potential, (2) general-purpose FMs can match medical-specific FMs if properly adapted, and (3) 2D-based methods surpass 3D architectures for 3D classification. For the first time, we demonstrate the feasibility of achieving state-of-the-art performance across diverse applications using a single scalable framework (including 1st place in the VLM3D challenge), eliminating the need for separate task-specific models.
VIViT: Variable-Input Vision Transformer Framework for 3D MR Image Segmentation
May 13, 2025Abstract:Self-supervised pretrain techniques have been widely used to improve the downstream tasks' performance. However, real-world magnetic resonance (MR) studies usually consist of different sets of contrasts due to different acquisition protocols, which poses challenges for the current deep learning methods on large-scale pretrain and different downstream tasks with different input requirements, since these methods typically require a fixed set of input modalities or, contrasts. To address this challenge, we propose variable-input ViT (VIViT), a transformer-based framework designed for self-supervised pretraining and segmentation finetuning for variable contrasts in each study. With this ability, our approach can maximize the data availability in pretrain, and can transfer the learned knowledge from pretrain to downstream tasks despite variations in input requirements. We validate our method on brain infarct and brain tumor segmentation, where our method outperforms current CNN and ViT-based models with a mean Dice score of 0.624 and 0.883 respectively. These results highlight the efficacy of our design for better adaptability and performance on tasks with real-world heterogeneous MR data.
Multi-Plane Vision Transformer for Hemorrhage Classification Using Axial and Sagittal MRI Data
May 12, 2025Abstract:Identifying brain hemorrhages from magnetic resonance imaging (MRI) is a critical task for healthcare professionals. The diverse nature of MRI acquisitions with varying contrasts and orientation introduce complexity in identifying hemorrhage using neural networks. For acquisitions with varying orientations, traditional methods often involve resampling images to a fixed plane, which can lead to information loss. To address this, we propose a 3D multi-plane vision transformer (MP-ViT) for hemorrhage classification with varying orientation data. It employs two separate transformer encoders for axial and sagittal contrasts, using cross-attention to integrate information across orientations. MP-ViT also includes a modality indication vector to provide missing contrast information to the model. The effectiveness of the proposed model is demonstrated with extensive experiments on real world clinical dataset consists of 10,084 training, 1,289 validation and 1,496 test subjects. MP-ViT achieved substantial improvement in area under the curve (AUC), outperforming the vision transformer (ViT) by 5.5% and CNN-based architectures by 1.8%. These results highlight the potential of MP-ViT in improving performance for hemorrhage detection when different orientation contrasts are needed.
AdaViT: Adaptive Vision Transformer for Flexible Pretrain and Finetune with Variable 3D Medical Image Modalities
Apr 04, 2025



Abstract:Pretrain techniques, whether supervised or self-supervised, are widely used in deep learning to enhance model performance. In real-world clinical scenarios, different sets of magnetic resonance (MR) contrasts are often acquired for different subjects/cases, creating challenges for deep learning models assuming consistent input modalities among all the cases and between pretrain and finetune. Existing methods struggle to maintain performance when there is an input modality/contrast set mismatch with the pretrained model, often resulting in degraded accuracy. We propose an adaptive Vision Transformer (AdaViT) framework capable of handling variable set of input modalities for each case. We utilize a dynamic tokenizer to encode different input image modalities to tokens and take advantage of the characteristics of the transformer to build attention mechanism across variable length of tokens. Through extensive experiments, we demonstrate that this architecture effectively transfers supervised pretrained models to new datasets with different input modality/contrast sets, resulting in superior performance on zero-shot testing, few-shot finetuning, and backward transferring in brain infarct and brain tumor segmentation tasks. Additionally, for self-supervised pretrain, the proposed method is able to maximize the pretrain data and facilitate transferring to diverse downstream tasks with variable sets of input modalities.
SegResMamba: An Efficient Architecture for 3D Medical Image Segmentation
Mar 10, 2025Abstract:The Transformer architecture has opened a new paradigm in the domain of deep learning with its ability to model long-range dependencies and capture global context and has outpaced the traditional Convolution Neural Networks (CNNs) in many aspects. However, applying Transformer models to 3D medical image datasets presents significant challenges due to their high training time, and memory requirements, which not only hinder scalability but also contribute to elevated CO$_2$ footprint. This has led to an exploration of alternative models that can maintain or even improve performance while being more efficient and environmentally sustainable. Recent advancements in Structured State Space Models (SSMs) effectively address some of the inherent limitations of Transformers, particularly their high memory and computational demands. Inspired by these advancements, we propose an efficient 3D segmentation model for medical imaging called SegResMamba, designed to reduce computation complexity, memory usage, training time, and environmental impact while maintaining high performance. Our model uses less than half the memory during training compared to other state-of-the-art (SOTA) architectures, achieving comparable performance with significantly reduced resource demands.
Self Pre-training with Adaptive Mask Autoencoders for Variable-Contrast 3D Medical Imaging
Jan 15, 2025Abstract:The Masked Autoencoder (MAE) has recently demonstrated effectiveness in pre-training Vision Transformers (ViT) for analyzing natural images. By reconstructing complete images from partially masked inputs, the ViT encoder gathers contextual information to predict the missing regions. This capability to aggregate context is especially important in medical imaging, where anatomical structures are functionally and mechanically linked to surrounding regions. However, current methods do not consider variations in the number of input images, which is typically the case in real-world Magnetic Resonance (MR) studies. To address this limitation, we propose a 3D Adaptive Masked Autoencoders (AMAE) architecture that accommodates a variable number of 3D input contrasts per subject. A magnetic resonance imaging (MRI) dataset of 45,364 subjects was used for pretraining and a subset of 1648 training, 193 validation and 215 test subjects were used for finetuning. The performance demonstrates that self pre-training of this adaptive masked autoencoders can enhance the infarct segmentation performance by 2.8%-3.7% for ViT-based segmentation models.
Bayesian convolutional neural network based MRI brain extraction on nonhuman primates
May 18, 2020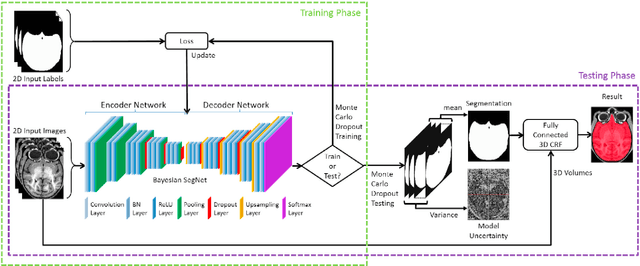
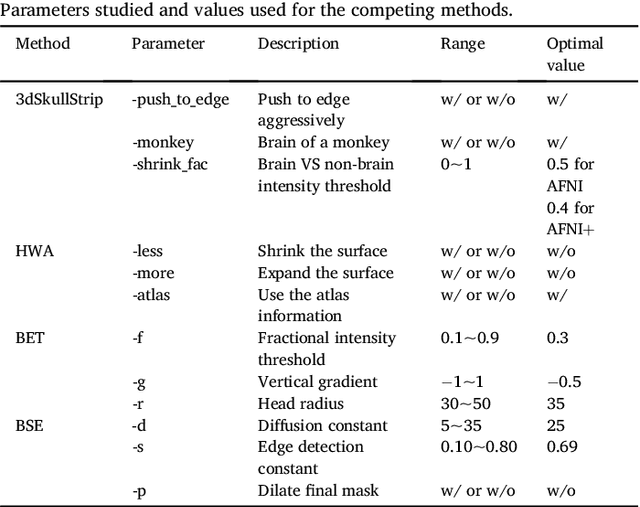
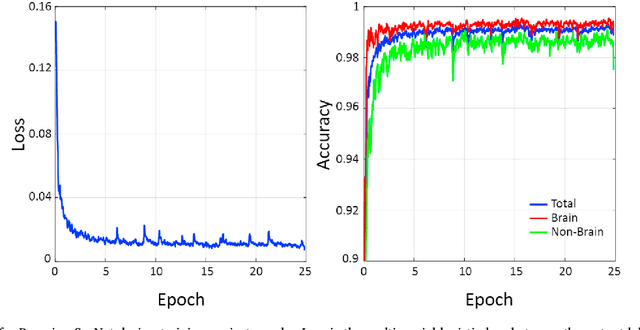
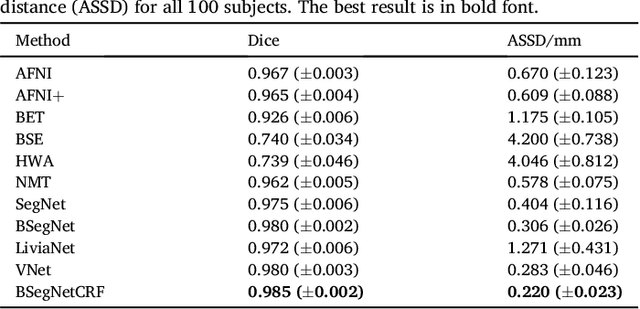
Abstract:Brain extraction or skull stripping of magnetic resonance images (MRI) is an essential step in neuroimaging studies, the accuracy of which can severely affect subsequent image processing procedures. Current automatic brain extraction methods demonstrate good results on human brains, but are often far from satisfactory on nonhuman primates, which are a necessary part of neuroscience research. To overcome the challenges of brain extraction in nonhuman primates, we propose a fully-automated brain extraction pipeline combining deep Bayesian convolutional neural network (CNN) and fully connected three-dimensional (3D) conditional random field (CRF). The deep Bayesian CNN, Bayesian SegNet, is used as the core segmentation engine. As a probabilistic network, it is not only able to perform accurate high-resolution pixel-wise brain segmentation, but also capable of measuring the model uncertainty by Monte Carlo sampling with dropout in the testing stage. Then, fully connected 3D CRF is used to refine the probability result from Bayesian SegNet in the whole 3D context of the brain volume. The proposed method was evaluated with a manually brain-extracted dataset comprising T1w images of 100 nonhuman primates. Our method outperforms six popular publicly available brain extraction packages and three well-established deep learning based methods with a mean Dice coefficient of 0.985 and a mean average symmetric surface distance of 0.220 mm. A better performance against all the compared methods was verified by statistical tests (all p-values<10-4, two-sided, Bonferroni corrected). The maximum uncertainty of the model on nonhuman primate brain extraction has a mean value of 0.116 across all the 100 subjects...
* 37 pages, 14 figures
Deep Learning and Bayesian Deep Learning Based Gender Prediction in Multi-Scale Brain Functional Connectivity
May 18, 2020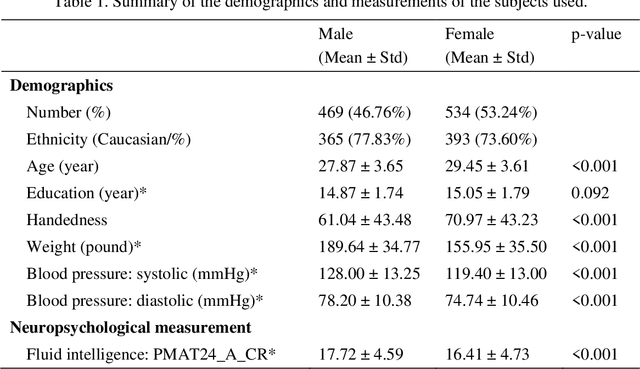
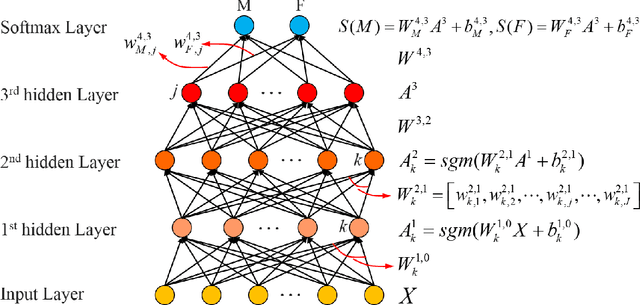
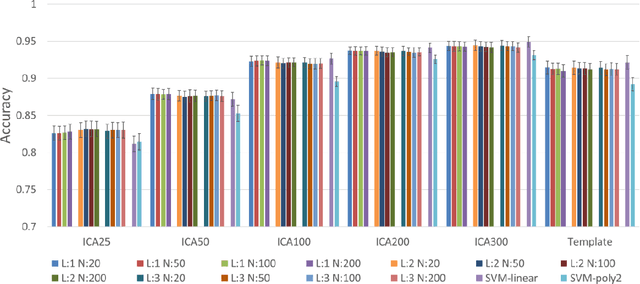
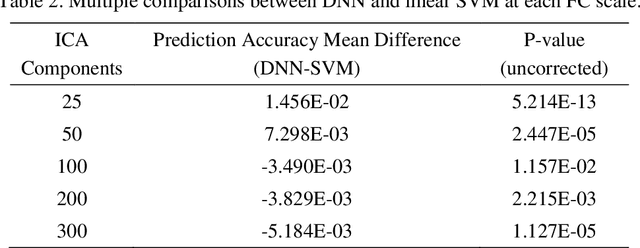
Abstract:Brain gender differences have been known for a long time and are the possible reason for many psychological, psychiatric and behavioral differences between males and females. Predicting genders from brain functional connectivity (FC) can build the relationship between brain activities and gender, and extracting important gender related FC features from the prediction model offers a way to investigate the brain gender difference. Current predictive models applied to gender prediction demonstrate good accuracies, but usually extract individual functional connections instead of connectivity patterns in the whole connectivity matrix as features. In addition, current models often omit the effect of the input brain FC scale on prediction and cannot give any model uncertainty information. Hence, in this study we propose to predict gender from multiple scales of brain FC with deep learning, which can extract full FC patterns as features. We further develop the understanding of the feature extraction mechanism in deep neural network (DNN) and propose a DNN feature ranking method to extract the highly important features based on their contributions to the prediction. Moreover, we apply Bayesian deep learning to the brain FC gender prediction, which as a probabilistic model can not only make accurate predictions but also generate model uncertainty for each prediction. Experiments were done on the high-quality Human Connectome Project S1200 release dataset comprising the resting state functional MRI data of 1003 healthy adults. First, DNN reaches 83.0%, 87.6%, 92.0%, 93.5% and 94.1% accuracies respectively with the FC input derived from 25, 50, 100, 200, 300 independent component analysis (ICA) components. DNN outperforms the conventional machine learning methods on the 25-ICA-component scale FC, but the linear machine learning method catches up as the number of ICA components increases...
Accurate Automatic Segmentation of Amygdala Subnuclei and Modeling of Uncertainty via Bayesian Fully Convolutional Neural Network
Feb 19, 2019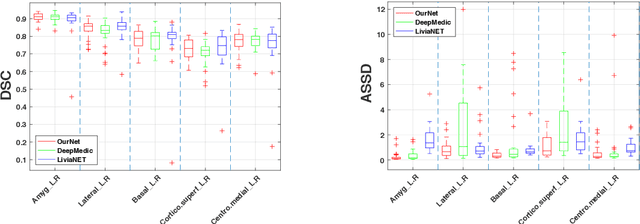
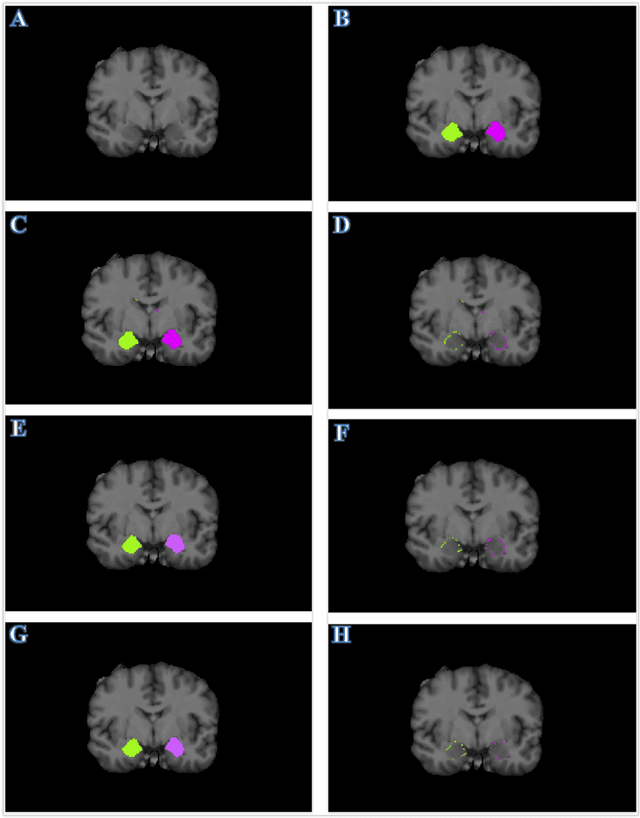
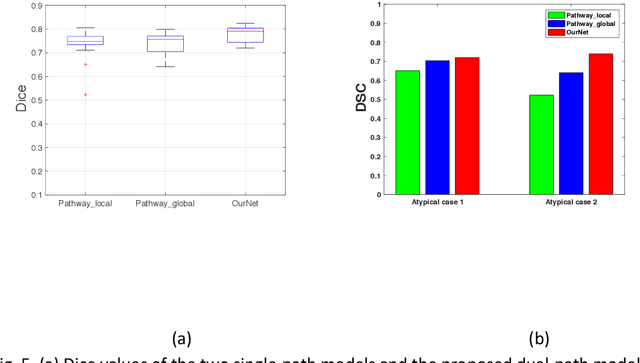
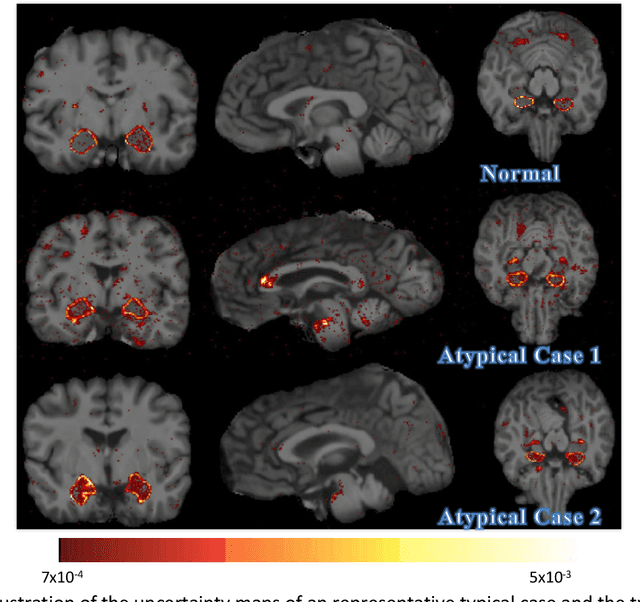
Abstract:Recent advances in deep learning have improved the segmentation accuracy of subcortical brain structures, which would be useful in neuroimaging studies of many neurological disorders. However, most of the previous deep learning work does not investigate the specific difficulties that exist in segmenting extremely small but important brain regions such as the amygdala and its subregions. To tackle this challenging task, a novel 3D Bayesian fully convolutional neural network was developed to apply a dilated dualpathway approach that retains fine details and utilizes both local and more global contextual information to automatically segment the amygdala and its subregions at high precision. The proposed method provides insights on network design and sampling strategy that target segmentations of small 3D structures. In particular, this study confirms that a large context, enabled by a large field of view, is beneficial for segmenting small objects; furthermore, precise contextual information enabled by dilated convolutions allows for better boundary localization, which is critical for examining the morphology of the structure. In addition, it is demonstrated that the uncertainty information estimated from our network may be leveraged to identify atypicality in data. Our method was compared with two state-of-the-art deep learning models and a traditional multi-atlas approach, and exhibited excellent performance as measured both by Dice overlap as well as average symmetric surface distance. To the best of our knowledge, this work is the first deep learning-based approach that targets the subregions of the amygdala.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge