Andrew L. Alexander
Stroke Lesion Segmentation using Multi-Stage Cross-Scale Attention
Jan 26, 2025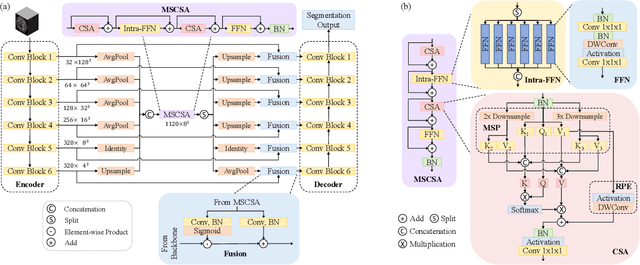
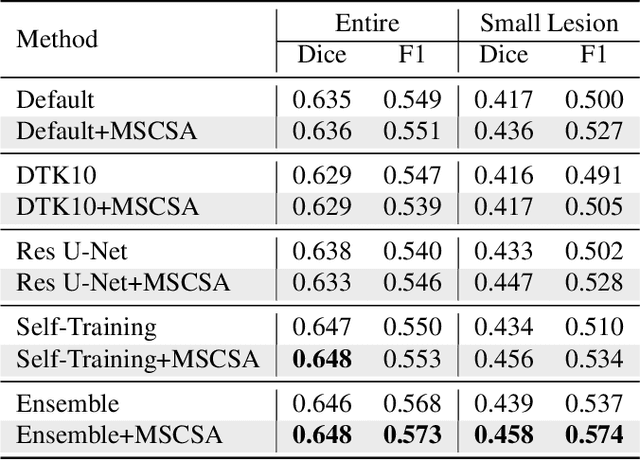
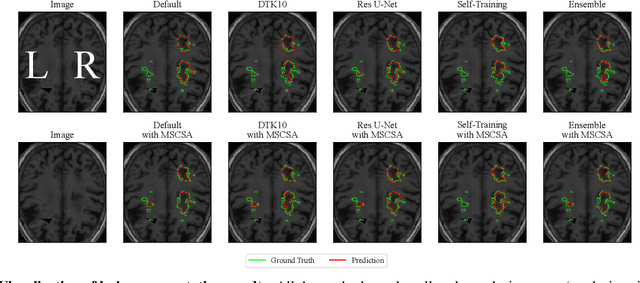
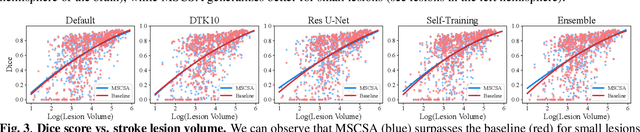
Abstract:Precise characterization of stroke lesions from MRI data has immense value in prognosticating clinical and cognitive outcomes following a stroke. Manual stroke lesion segmentation is time-consuming and requires the expertise of neurologists and neuroradiologists. Often, lesions are grossly characterized for their location and overall extent using bounding boxes without specific delineation of their boundaries. While such characterization provides some clinical value, to develop a precise mechanistic understanding of the impact of lesions on post-stroke vascular contributions to cognitive impairments and dementia (VCID), the stroke lesions need to be fully segmented with accurate boundaries. This work introduces the Multi-Stage Cross-Scale Attention (MSCSA) mechanism, applied to the U-Net family, to improve the mapping between brain structural features and lesions of varying sizes. Using the Anatomical Tracings of Lesions After Stroke (ATLAS) v2.0 dataset, MSCSA outperforms all baseline methods in both Dice and F1 scores on a subset focusing on small lesions, while maintaining competitive performance across the entire dataset. Notably, the ensemble strategy incorporating MSCSA achieves the highest scores for Dice and F1 on both the full dataset and the small lesion subset. These results demonstrate the effectiveness of MSCSA in segmenting small lesions and highlight its robustness across different training schemes for large stroke lesions. Our code is available at: https://github.com/nadluru/StrokeLesSeg.
Segmenting Small Stroke Lesions with Novel Labeling Strategies
Aug 06, 2024Abstract:Deep neural networks have demonstrated exceptional efficacy in stroke lesion segmentation. However, the delineation of small lesions, critical for stroke diagnosis, remains a challenge. In this study, we propose two straightforward yet powerful approaches that can be seamlessly integrated into a variety of networks: Multi-Size Labeling (MSL) and Distance-Based Labeling (DBL), with the aim of enhancing the segmentation accuracy of small lesions. MSL divides lesion masks into various categories based on lesion volume while DBL emphasizes the lesion boundaries. Experimental evaluations on the Anatomical Tracings of Lesions After Stroke (ATLAS) v2.0 dataset showcase that an ensemble of MSL and DBL achieves consistently better or equal performance on recall (3.6% and 3.7%), F1 (2.4% and 1.5%), and Dice scores (1.3% and 0.0%) compared to the top-1 winner of the 2022 MICCAI ATLAS Challenge on both the subset only containing small lesions and the entire dataset, respectively. Notably, on the mini-lesion subset, a single MSL model surpasses the previous best ensemble strategy, with enhancements of 1.0% and 0.3% on F1 and Dice scores, respectively. Our code is available at: https://github.com/nadluru/StrokeLesSeg.
Accurate Automatic Segmentation of Amygdala Subnuclei and Modeling of Uncertainty via Bayesian Fully Convolutional Neural Network
Feb 19, 2019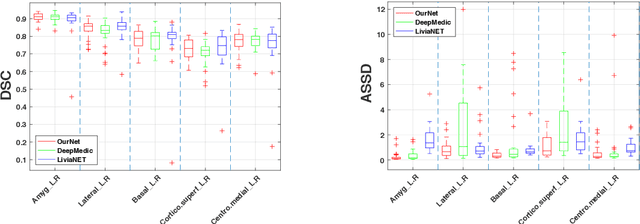
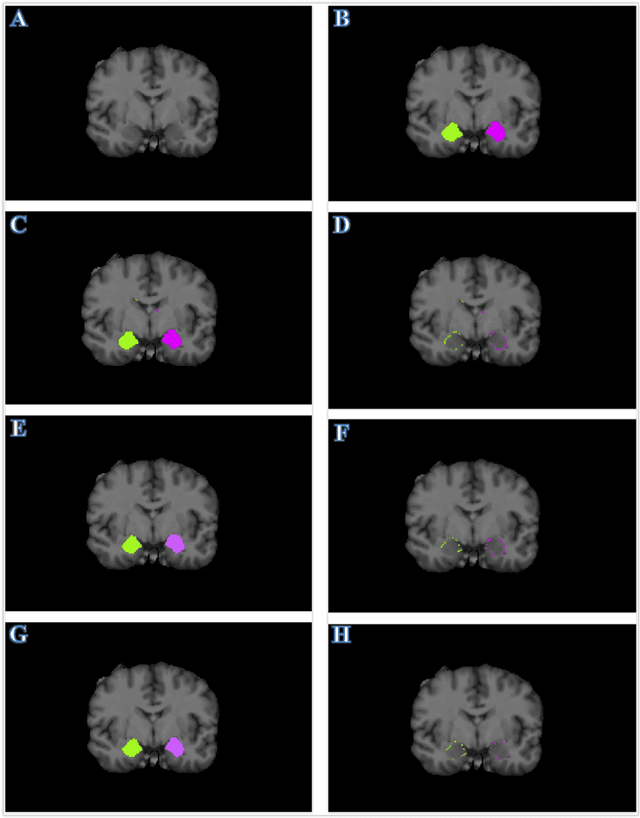
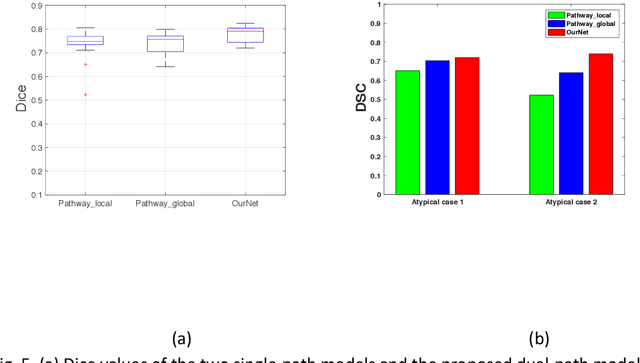
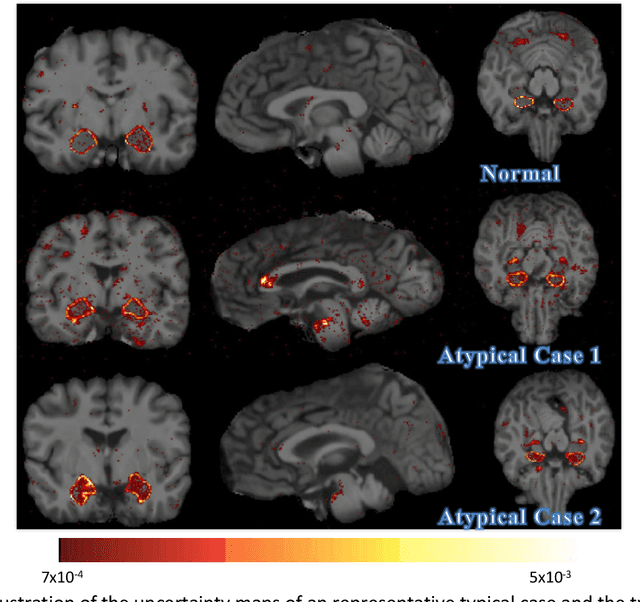
Abstract:Recent advances in deep learning have improved the segmentation accuracy of subcortical brain structures, which would be useful in neuroimaging studies of many neurological disorders. However, most of the previous deep learning work does not investigate the specific difficulties that exist in segmenting extremely small but important brain regions such as the amygdala and its subregions. To tackle this challenging task, a novel 3D Bayesian fully convolutional neural network was developed to apply a dilated dualpathway approach that retains fine details and utilizes both local and more global contextual information to automatically segment the amygdala and its subregions at high precision. The proposed method provides insights on network design and sampling strategy that target segmentations of small 3D structures. In particular, this study confirms that a large context, enabled by a large field of view, is beneficial for segmenting small objects; furthermore, precise contextual information enabled by dilated convolutions allows for better boundary localization, which is critical for examining the morphology of the structure. In addition, it is demonstrated that the uncertainty information estimated from our network may be leveraged to identify atypicality in data. Our method was compared with two state-of-the-art deep learning models and a traditional multi-atlas approach, and exhibited excellent performance as measured both by Dice overlap as well as average symmetric surface distance. To the best of our knowledge, this work is the first deep learning-based approach that targets the subregions of the amygdala.
Diffeomorphic Metric Mapping and Probabilistic Atlas Generation of Hybrid Diffusion Imaging based on BFOR Signal Basis
Sep 25, 2013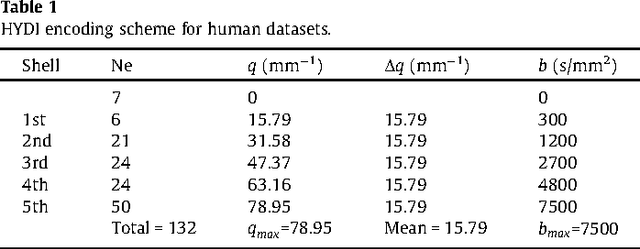
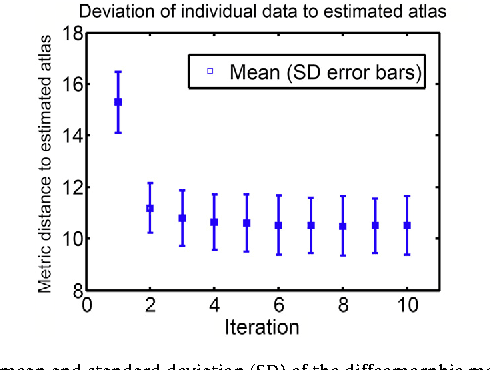
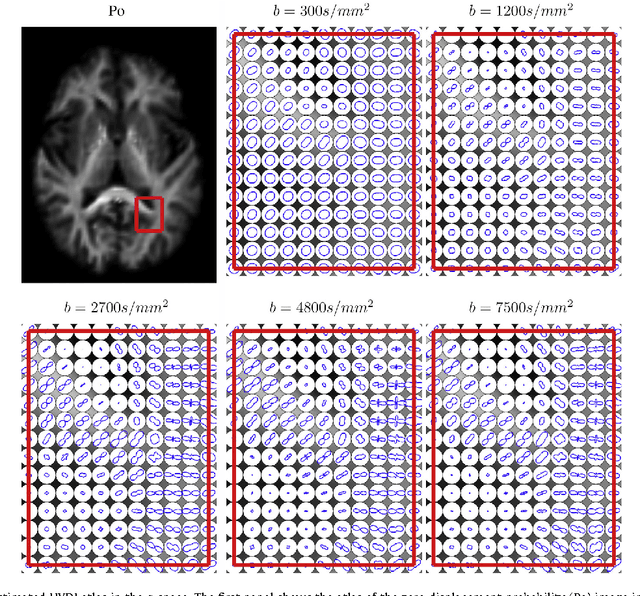
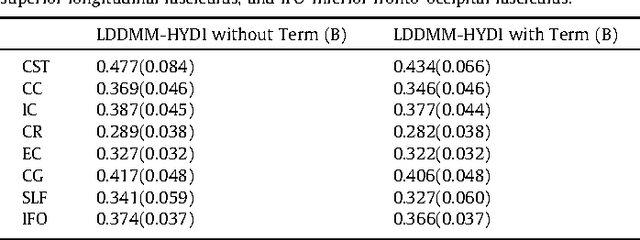
Abstract:We propose a large deformation diffeomorphic metric mapping algorithm to align multiple b-value diffusion weighted imaging (mDWI) data, specifically acquired via hybrid diffusion imaging (HYDI), denoted as LDDMM-HYDI. We then propose a Bayesian model for estimating the white matter atlas from HYDIs. We adopt the work given in Hosseinbor et al. (2012) and represent the q-space diffusion signal with the Bessel Fourier orientation reconstruction (BFOR) signal basis. The BFOR framework provides the representation of mDWI in the q-space and thus reduces memory requirement. In addition, since the BFOR signal basis is orthonormal, the L2 norm that quantifies the differences in the q-space signals of any two mDWI datasets can be easily computed as the sum of the squared differences in the BFOR expansion coefficients. In this work, we show that the reorientation of the $q$-space signal due to spatial transformation can be easily defined on the BFOR signal basis. We incorporate the BFOR signal basis into the LDDMM framework and derive the gradient descent algorithm for LDDMM-HYDI with explicit orientation optimization. Additionally, we extend the previous Bayesian atlas estimation framework for scalar-valued images to HYDIs and derive the expectation-maximization algorithm for solving the HYDI atlas estimation problem. Using real HYDI datasets, we show the Bayesian model generates the white matter atlas with anatomical details. Moreover, we show that it is important to consider the variation of mDWI reorientation due to a small change in diffeomorphic transformation in the LDDMM-HYDI optimization and to incorporate the full information of HYDI for aligning mDWI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge