Gyujoon Hwang
from the iSTAGING consortium, for the ADNI
Dimensional Neuroimaging Endophenotypes: Neurobiological Representations of Disease Heterogeneity Through Machine Learning
Jan 17, 2024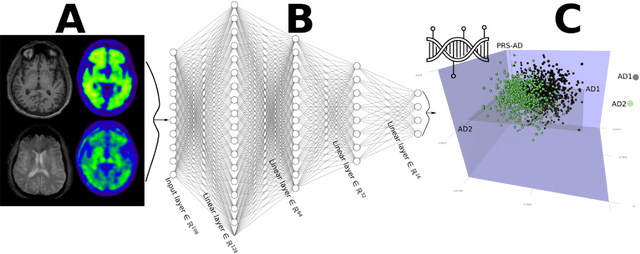
Abstract:Machine learning has been increasingly used to obtain individualized neuroimaging signatures for disease diagnosis, prognosis, and response to treatment in neuropsychiatric and neurodegenerative disorders. Therefore, it has contributed to a better understanding of disease heterogeneity by identifying disease subtypes that present significant differences in various brain phenotypic measures. In this review, we first present a systematic literature overview of studies using machine learning and multimodal MRI to unravel disease heterogeneity in various neuropsychiatric and neurodegenerative disorders, including Alzheimer disease, schizophrenia, major depressive disorder, autism spectrum disorder, multiple sclerosis, as well as their potential in transdiagnostic settings. Subsequently, we summarize relevant machine learning methodologies and discuss an emerging paradigm which we call dimensional neuroimaging endophenotype (DNE). DNE dissects the neurobiological heterogeneity of neuropsychiatric and neurodegenerative disorders into a low dimensional yet informative, quantitative brain phenotypic representation, serving as a robust intermediate phenotype (i.e., endophenotype) largely reflecting underlying genetics and etiology. Finally, we discuss the potential clinical implications of the current findings and envision future research avenues.
Subtyping brain diseases from imaging data
Feb 16, 2022

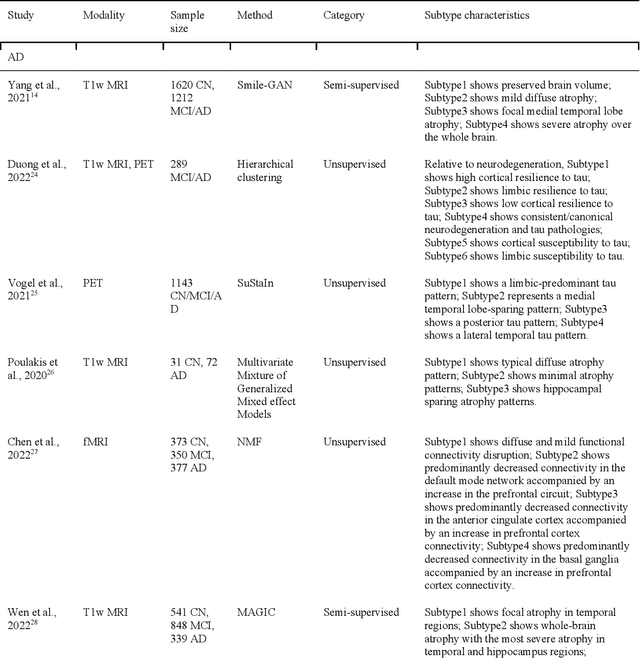
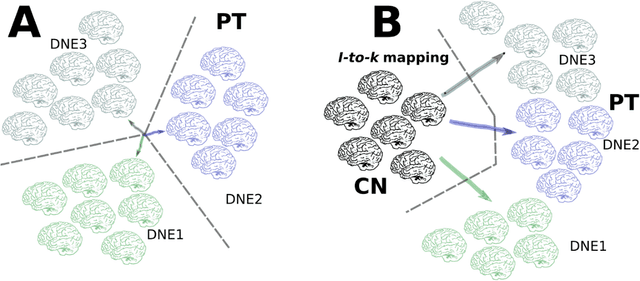
Abstract:The imaging community has increasingly adopted machine learning (ML) methods to provide individualized imaging signatures related to disease diagnosis, prognosis, and response to treatment. Clinical neuroscience and cancer imaging have been two areas in which ML has offered particular promise. However, many neurologic and neuropsychiatric diseases, as well as cancer, are often heterogeneous in terms of their clinical manifestations, neuroanatomical patterns or genetic underpinnings. Therefore, in such cases, seeking a single disease signature might be ineffectual in delivering individualized precision diagnostics. The current chapter focuses on ML methods, especially semi-supervised clustering, that seek disease subtypes using imaging data. Work from Alzheimer Disease and its prodromal stages, psychosis, depression, autism, and brain cancer are discussed. Our goal is to provide the readers with a broad overview in terms of methodology and clinical applications.
Disentangling Alzheimer's disease neurodegeneration from typical brain aging using machine learning
Sep 08, 2021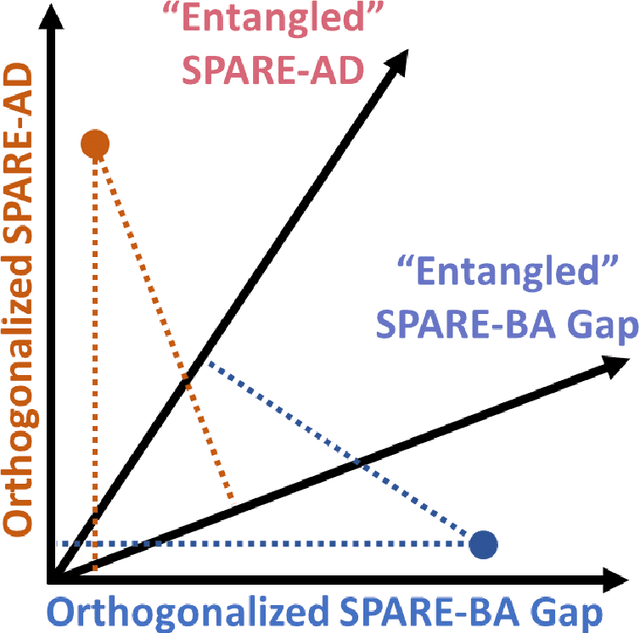
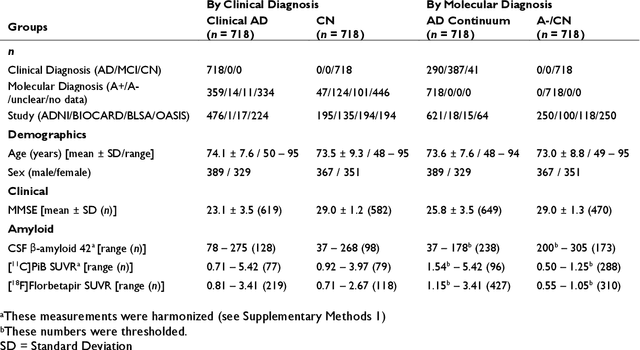
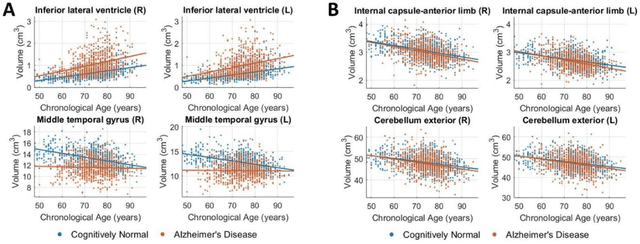
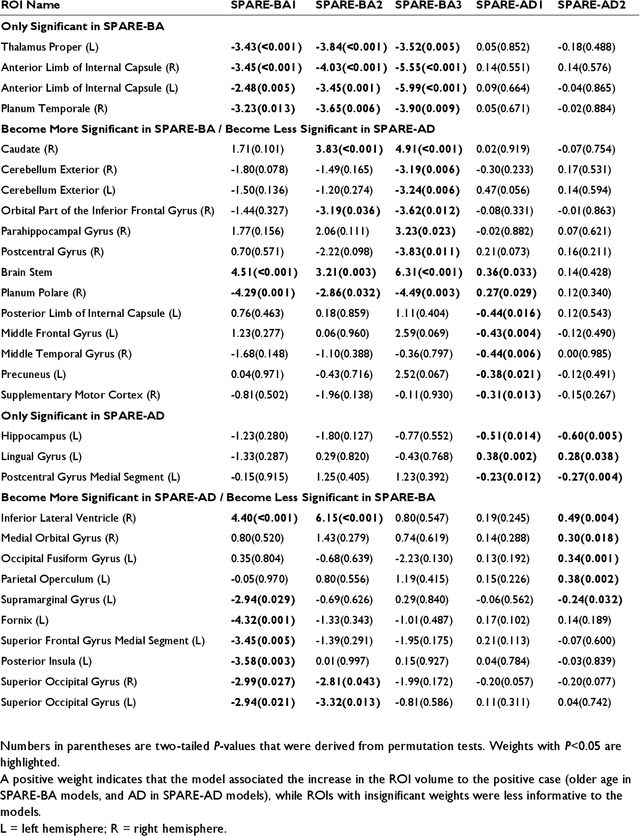
Abstract:Neuroimaging biomarkers that distinguish between typical brain aging and Alzheimer's disease (AD) are valuable for determining how much each contributes to cognitive decline. Machine learning models can derive multi-variate brain change patterns related to the two processes, including the SPARE-AD (Spatial Patterns of Atrophy for Recognition of Alzheimer's Disease) and SPARE-BA (of Brain Aging) investigated herein. However, substantial overlap between brain regions affected in the two processes confounds measuring them independently. We present a methodology toward disentangling the two. T1-weighted MRI images of 4,054 participants (48-95 years) with AD, mild cognitive impairment (MCI), or cognitively normal (CN) diagnoses from the iSTAGING (Imaging-based coordinate SysTem for AGIng and NeurodeGenerative diseases) consortium were analyzed. First, a subset of AD patients and CN adults were selected based purely on clinical diagnoses to train SPARE-BA1 (regression of age using CN individuals) and SPARE-AD1 (classification of CN versus AD). Second, analogous groups were selected based on clinical and molecular markers to train SPARE-BA2 and SPARE-AD2: amyloid-positive (A+) AD continuum group (consisting of A+AD, A+MCI, and A+ and tau-positive CN individuals) and amyloid-negative (A-) CN group. Finally, the combined group of the AD continuum and A-/CN individuals was used to train SPARE-BA3, with the intention to estimate brain age regardless of AD-related brain changes. Disentangled SPARE models derived brain patterns that were more specific to the two types of the brain changes. Correlation between the SPARE-BA and SPARE-AD was significantly reduced. Correlation of disentangled SPARE-AD was non-inferior to the molecular measurements and to the number of APOE4 alleles, but was less to AD-related psychometric test scores, suggesting contribution of advanced brain aging to these scores.
Deep Learning and Bayesian Deep Learning Based Gender Prediction in Multi-Scale Brain Functional Connectivity
May 18, 2020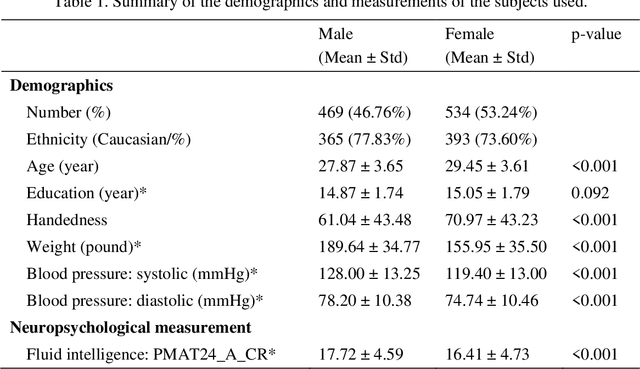
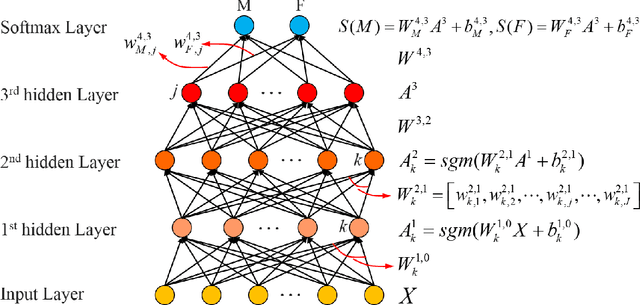
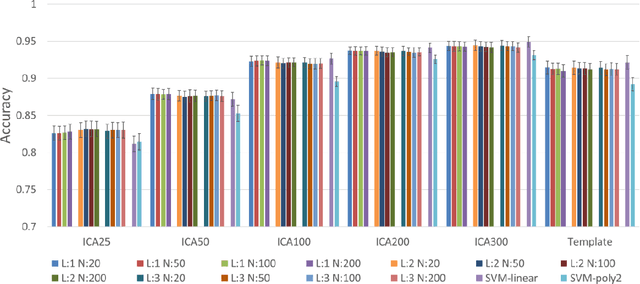
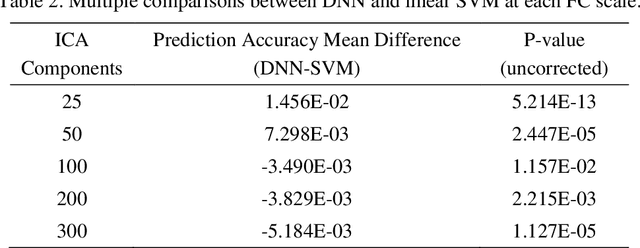
Abstract:Brain gender differences have been known for a long time and are the possible reason for many psychological, psychiatric and behavioral differences between males and females. Predicting genders from brain functional connectivity (FC) can build the relationship between brain activities and gender, and extracting important gender related FC features from the prediction model offers a way to investigate the brain gender difference. Current predictive models applied to gender prediction demonstrate good accuracies, but usually extract individual functional connections instead of connectivity patterns in the whole connectivity matrix as features. In addition, current models often omit the effect of the input brain FC scale on prediction and cannot give any model uncertainty information. Hence, in this study we propose to predict gender from multiple scales of brain FC with deep learning, which can extract full FC patterns as features. We further develop the understanding of the feature extraction mechanism in deep neural network (DNN) and propose a DNN feature ranking method to extract the highly important features based on their contributions to the prediction. Moreover, we apply Bayesian deep learning to the brain FC gender prediction, which as a probabilistic model can not only make accurate predictions but also generate model uncertainty for each prediction. Experiments were done on the high-quality Human Connectome Project S1200 release dataset comprising the resting state functional MRI data of 1003 healthy adults. First, DNN reaches 83.0%, 87.6%, 92.0%, 93.5% and 94.1% accuracies respectively with the FC input derived from 25, 50, 100, 200, 300 independent component analysis (ICA) components. DNN outperforms the conventional machine learning methods on the 25-ICA-component scale FC, but the linear machine learning method catches up as the number of ICA components increases...
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge