Zhijian Yang
Can NeRFs See without Cameras?
May 28, 2025Abstract:Neural Radiance Fields (NeRFs) have been remarkably successful at synthesizing novel views of 3D scenes by optimizing a volumetric scene function. This scene function models how optical rays bring color information from a 3D object to the camera pixels. Radio frequency (RF) or audio signals can also be viewed as a vehicle for delivering information about the environment to a sensor. However, unlike camera pixels, an RF/audio sensor receives a mixture of signals that contain many environmental reflections (also called "multipath"). Is it still possible to infer the environment using such multipath signals? We show that with redesign, NeRFs can be taught to learn from multipath signals, and thereby "see" the environment. As a grounding application, we aim to infer the indoor floorplan of a home from sparse WiFi measurements made at multiple locations inside the home. Although a difficult inverse problem, our implicitly learnt floorplans look promising, and enables forward applications, such as indoor signal prediction and basic ray tracing.
Enhancing SAM with Efficient Prompting and Preference Optimization for Semi-supervised Medical Image Segmentation
Mar 06, 2025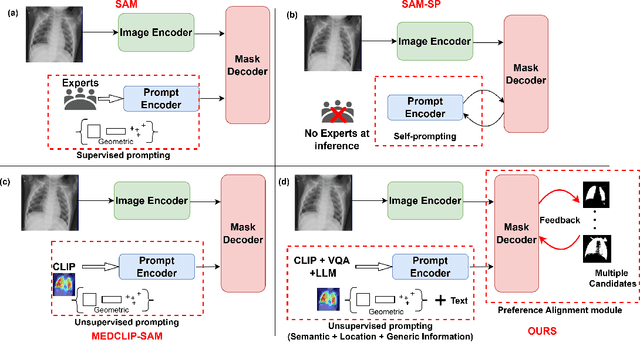
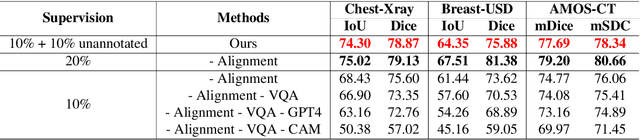
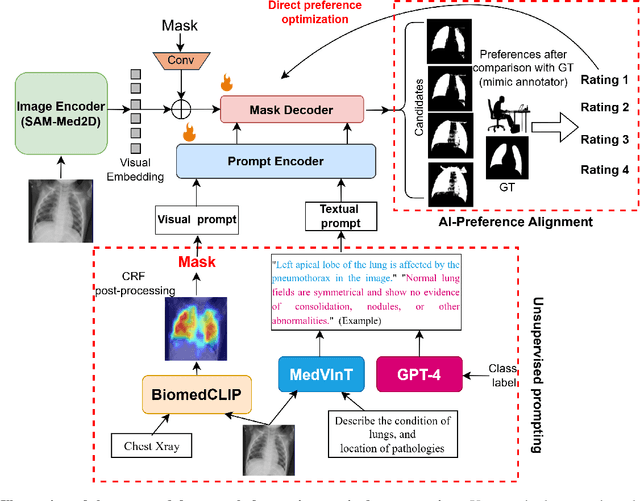
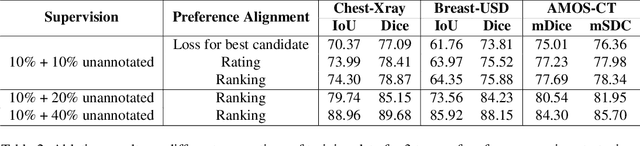
Abstract:Foundational models such as the Segment Anything Model (SAM) are gaining traction in medical imaging segmentation, supporting multiple downstream tasks. However, such models are supervised in nature, still relying on large annotated datasets or prompts supplied by experts. Conventional techniques such as active learning to alleviate such limitations are limited in scope and still necessitate continuous human involvement and complex domain knowledge for label refinement or establishing reward ground truth. To address these challenges, we propose an enhanced Segment Anything Model (SAM) framework that utilizes annotation-efficient prompts generated in a fully unsupervised fashion, while still capturing essential semantic, location, and shape information through contrastive language-image pretraining and visual question answering. We adopt the direct preference optimization technique to design an optimal policy that enables the model to generate high-fidelity segmentations with simple ratings or rankings provided by a virtual annotator simulating the human annotation process. State-of-the-art performance of our framework in tasks such as lung segmentation, breast tumor segmentation, and organ segmentation across various modalities, including X-ray, ultrasound, and abdominal CT, justifies its effectiveness in low-annotation data scenarios.
NeuroSynth: MRI-Derived Neuroanatomical Generative Models and Associated Dataset of 18,000 Samples
Jul 17, 2024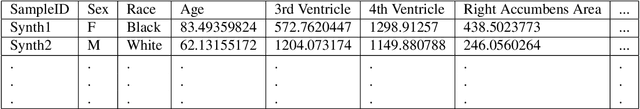
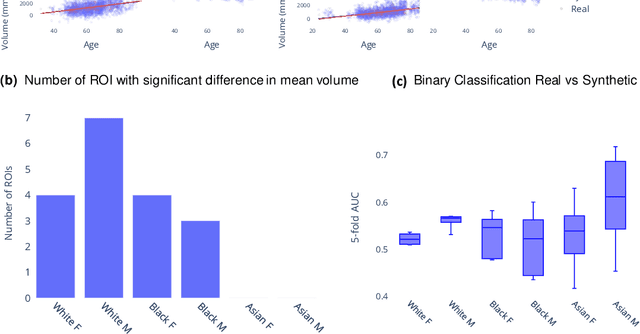
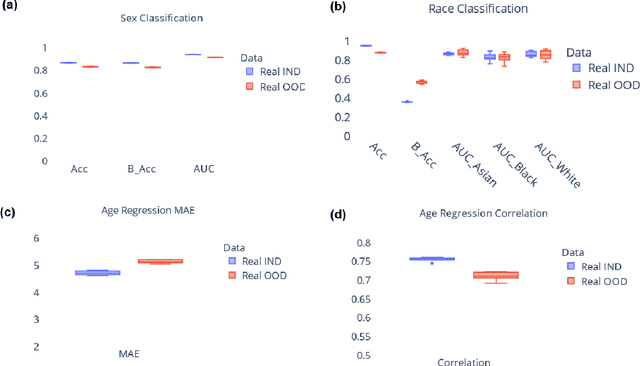
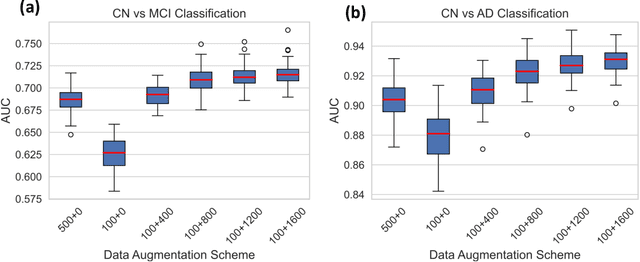
Abstract:Availability of large and diverse medical datasets is often challenged by privacy and data sharing restrictions. For successful application of machine learning techniques for disease diagnosis, prognosis, and precision medicine, large amounts of data are necessary for model building and optimization. To help overcome such limitations in the context of brain MRI, we present NeuroSynth: a collection of generative models of normative regional volumetric features derived from structural brain imaging. NeuroSynth models are trained on real brain imaging regional volumetric measures from the iSTAGING consortium, which encompasses over 40,000 MRI scans across 13 studies, incorporating covariates such as age, sex, and race. Leveraging NeuroSynth, we produce and offer 18,000 synthetic samples spanning the adult lifespan (ages 22-90 years), alongside the model's capability to generate unlimited data. Experimental results indicate that samples generated from NeuroSynth agree with the distributions obtained from real data. Most importantly, the generated normative data significantly enhance the accuracy of downstream machine learning models on tasks such as disease classification. Data and models are available at: https://huggingface.co/spaces/rongguangw/neuro-synth.
Dimensional Neuroimaging Endophenotypes: Neurobiological Representations of Disease Heterogeneity Through Machine Learning
Jan 17, 2024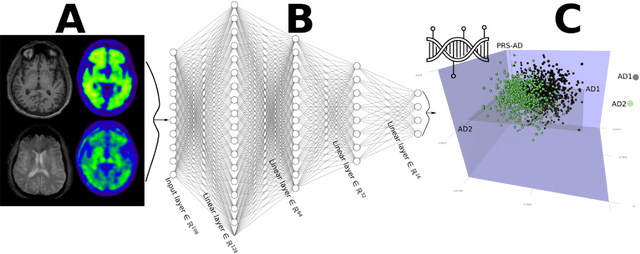
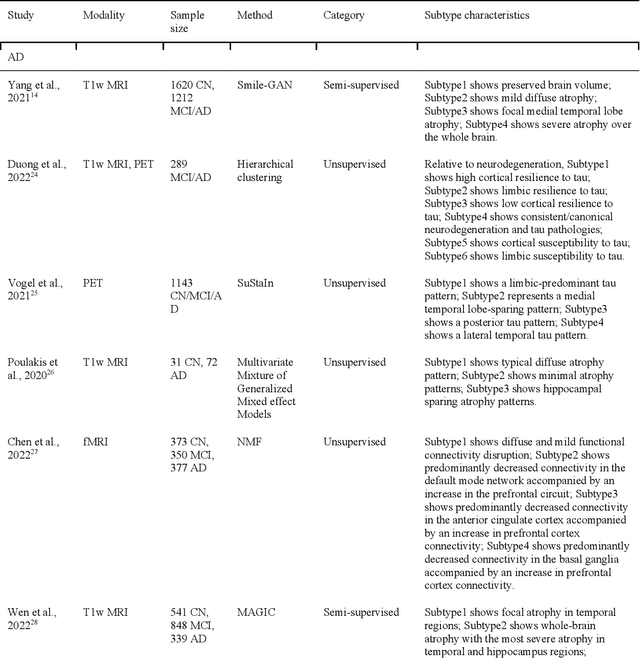
Abstract:Machine learning has been increasingly used to obtain individualized neuroimaging signatures for disease diagnosis, prognosis, and response to treatment in neuropsychiatric and neurodegenerative disorders. Therefore, it has contributed to a better understanding of disease heterogeneity by identifying disease subtypes that present significant differences in various brain phenotypic measures. In this review, we first present a systematic literature overview of studies using machine learning and multimodal MRI to unravel disease heterogeneity in various neuropsychiatric and neurodegenerative disorders, including Alzheimer disease, schizophrenia, major depressive disorder, autism spectrum disorder, multiple sclerosis, as well as their potential in transdiagnostic settings. Subsequently, we summarize relevant machine learning methodologies and discuss an emerging paradigm which we call dimensional neuroimaging endophenotype (DNE). DNE dissects the neurobiological heterogeneity of neuropsychiatric and neurodegenerative disorders into a low dimensional yet informative, quantitative brain phenotypic representation, serving as a robust intermediate phenotype (i.e., endophenotype) largely reflecting underlying genetics and etiology. Finally, we discuss the potential clinical implications of the current findings and envision future research avenues.
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023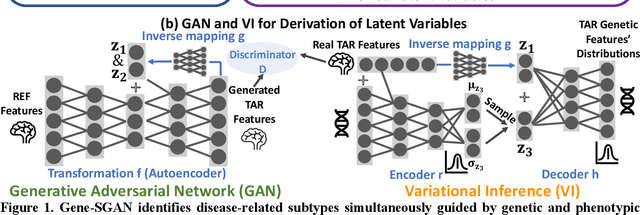
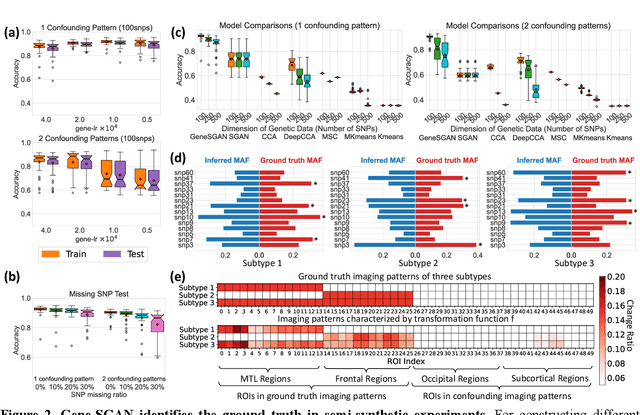
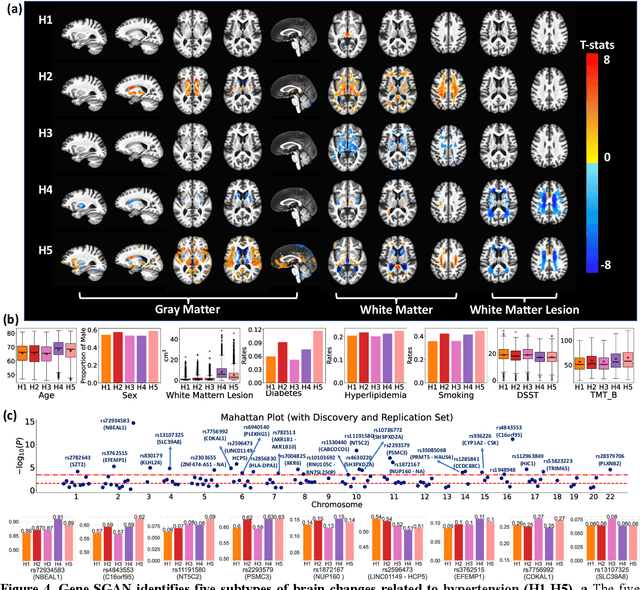
Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Applications of Generative Adversarial Networks in Neuroimaging and Clinical Neuroscience
Jun 14, 2022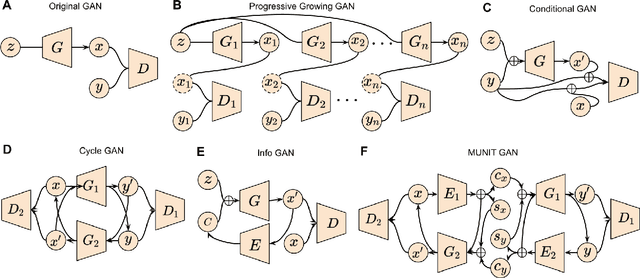
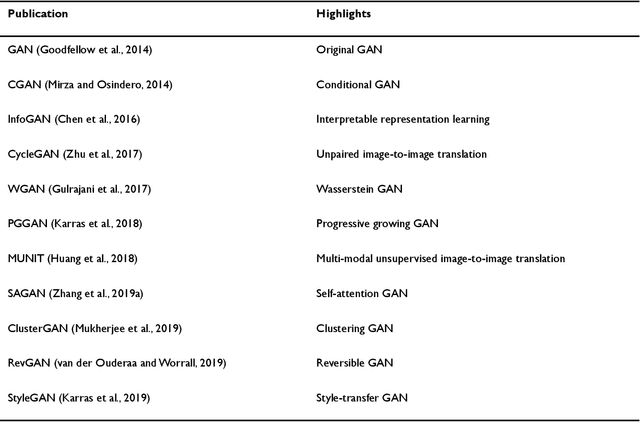
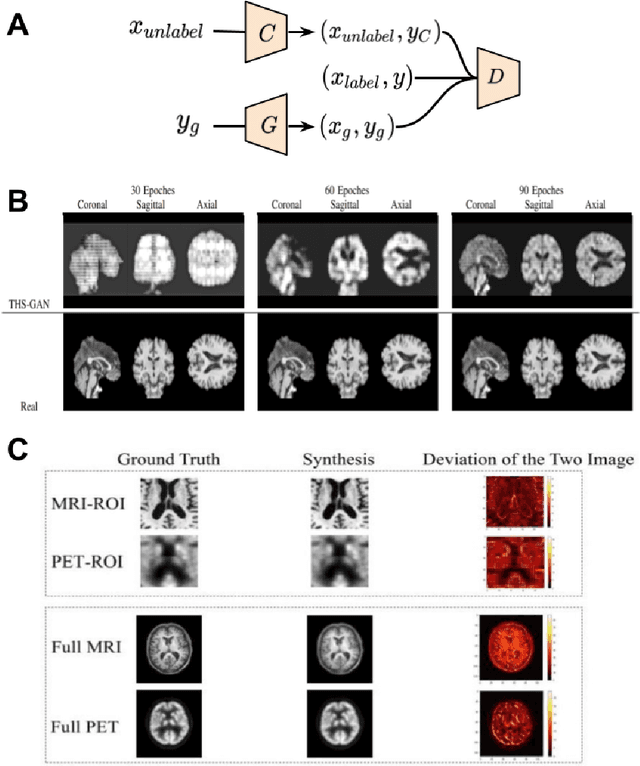
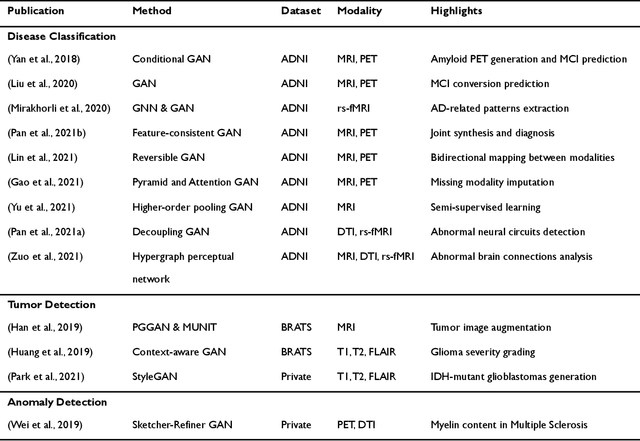
Abstract:Generative adversarial networks (GANs) are one powerful type of deep learning models that have been successfully utilized in numerous fields. They belong to a broader family called generative methods, which generate new data with a probabilistic model by learning sample distribution from real examples. In the clinical context, GANs have shown enhanced capabilities in capturing spatially complex, nonlinear, and potentially subtle disease effects compared to traditional generative methods. This review appraises the existing literature on the applications of GANs in imaging studies of various neurological conditions, including Alzheimer's disease, brain tumors, brain aging, and multiple sclerosis. We provide an intuitive explanation of various GAN methods for each application and further discuss the main challenges, open questions, and promising future directions of leveraging GANs in neuroimaging. We aim to bridge the gap between advanced deep learning methods and neurology research by highlighting how GANs can be leveraged to support clinical decision making and contribute to a better understanding of the structural and functional patterns of brain diseases.
Surreal-GAN:Semi-Supervised Representation Learning via GAN for uncovering heterogeneous disease-related imaging patterns
May 09, 2022



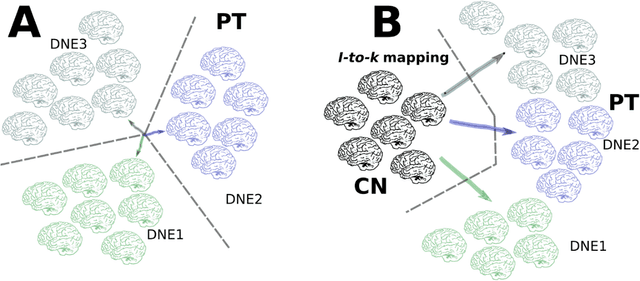
Abstract:A plethora of machine learning methods have been applied to imaging data, enabling the construction of clinically relevant imaging signatures of neurological and neuropsychiatric diseases. Oftentimes, such methods don't explicitly model the heterogeneity of disease effects, or approach it via nonlinear models that are not interpretable. Moreover, unsupervised methods may parse heterogeneity that is driven by nuisance confounding factors that affect brain structure or function, rather than heterogeneity relevant to a pathology of interest. On the other hand, semi-supervised clustering methods seek to derive a dichotomous subtype membership, ignoring the truth that disease heterogeneity spatially and temporally extends along a continuum. To address the aforementioned limitations, herein, we propose a novel method, termed Surreal-GAN (Semi-SUpeRvised ReprEsentAtion Learning via GAN). Using cross-sectional imaging data, Surreal-GAN dissects underlying disease-related heterogeneity under the principle of semi-supervised clustering (cluster mappings from normal control to patient), proposes a continuously dimensional representation, and infers the disease severity of patients at individual level along each dimension. The model first learns a transformation function from normal control (CN) domain to the patient (PT) domain with latent variables controlling transformation directions. An inverse mapping function together with regularization on function continuity, pattern orthogonality and monotonicity was also imposed to make sure that the transformation function captures necessarily meaningful imaging patterns with clinical significance. We first validated the model through extensive semi-synthetic experiments, and then demonstrate its potential in capturing biologically plausible imaging patterns in Alzheimer's disease (AD).
Subtyping brain diseases from imaging data
Feb 16, 2022

Abstract:The imaging community has increasingly adopted machine learning (ML) methods to provide individualized imaging signatures related to disease diagnosis, prognosis, and response to treatment. Clinical neuroscience and cancer imaging have been two areas in which ML has offered particular promise. However, many neurologic and neuropsychiatric diseases, as well as cancer, are often heterogeneous in terms of their clinical manifestations, neuroanatomical patterns or genetic underpinnings. Therefore, in such cases, seeking a single disease signature might be ineffectual in delivering individualized precision diagnostics. The current chapter focuses on ML methods, especially semi-supervised clustering, that seek disease subtypes using imaging data. Work from Alzheimer Disease and its prodromal stages, psychosis, depression, autism, and brain cancer are discussed. Our goal is to provide the readers with a broad overview in terms of methodology and clinical applications.
PoseKernelLifter: Metric Lifting of 3D Human Pose using Sound
Dec 03, 2021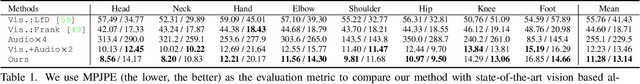
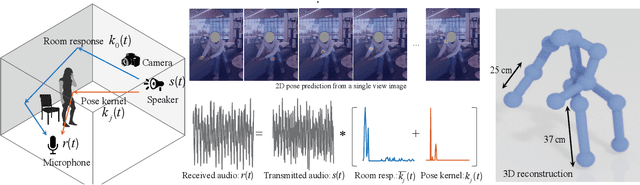
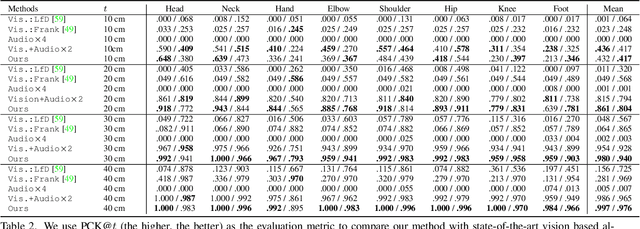
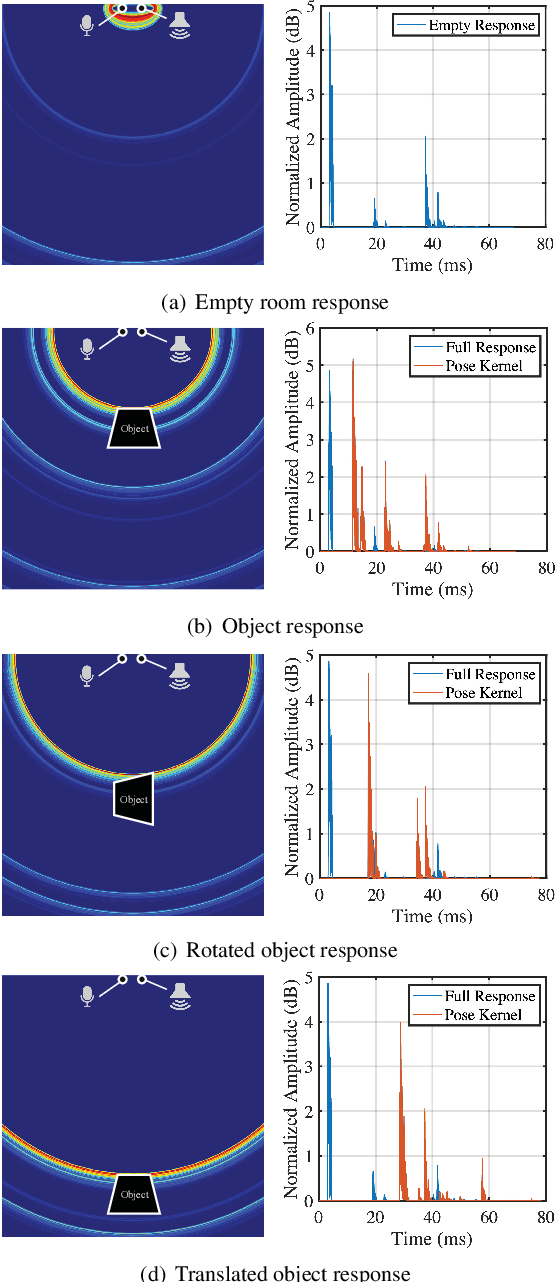
Abstract:Reconstructing the 3D pose of a person in metric scale from a single view image is a geometrically ill-posed problem. For example, we can not measure the exact distance of a person to the camera from a single view image without additional scene assumptions (e.g., known height). Existing learning based approaches circumvent this issue by reconstructing the 3D pose up to scale. However, there are many applications such as virtual telepresence, robotics, and augmented reality that require metric scale reconstruction. In this paper, we show that audio signals recorded along with an image, provide complementary information to reconstruct the metric 3D pose of the person. The key insight is that as the audio signals traverse across the 3D space, their interactions with the body provide metric information about the body's pose. Based on this insight, we introduce a time-invariant transfer function called pose kernel -- the impulse response of audio signals induced by the body pose. The main properties of the pose kernel are that (1) its envelope highly correlates with 3D pose, (2) the time response corresponds to arrival time, indicating the metric distance to the microphone, and (3) it is invariant to changes in the scene geometry configurations. Therefore, it is readily generalizable to unseen scenes. We design a multi-stage 3D CNN that fuses audio and visual signals and learns to reconstruct 3D pose in a metric scale. We show that our multi-modal method produces accurate metric reconstruction in real world scenes, which is not possible with state-of-the-art lifting approaches including parametric mesh regression and depth regression.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021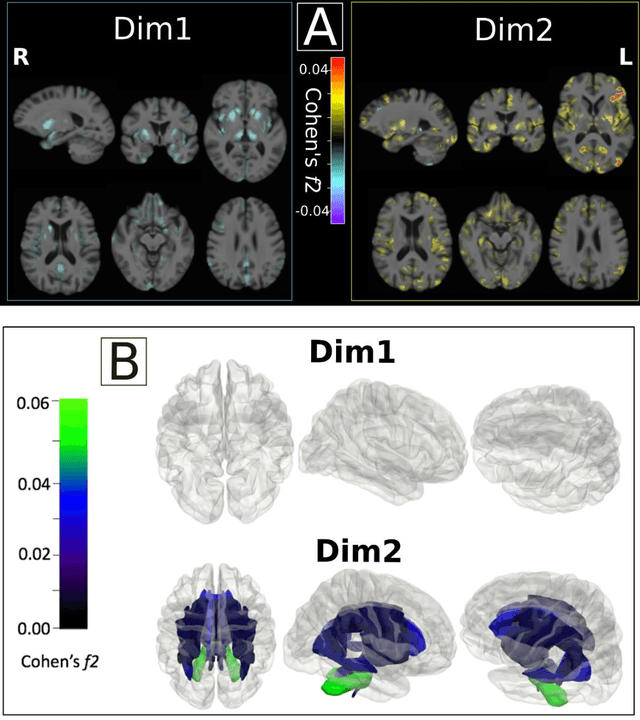
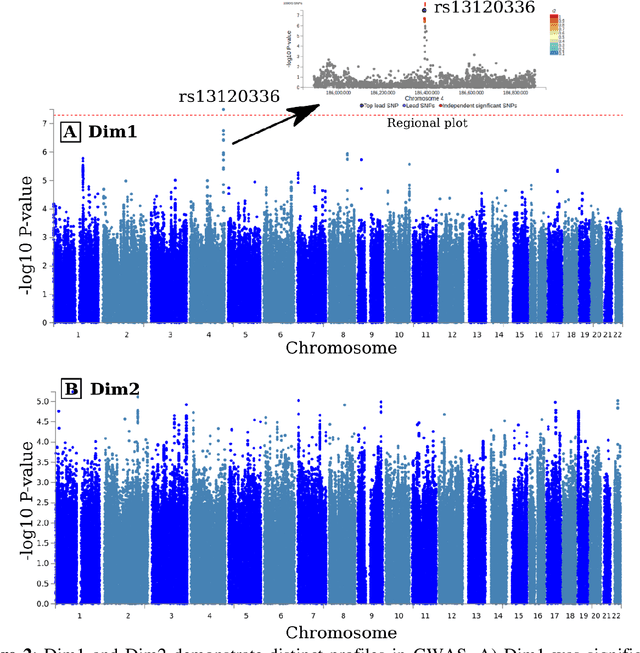
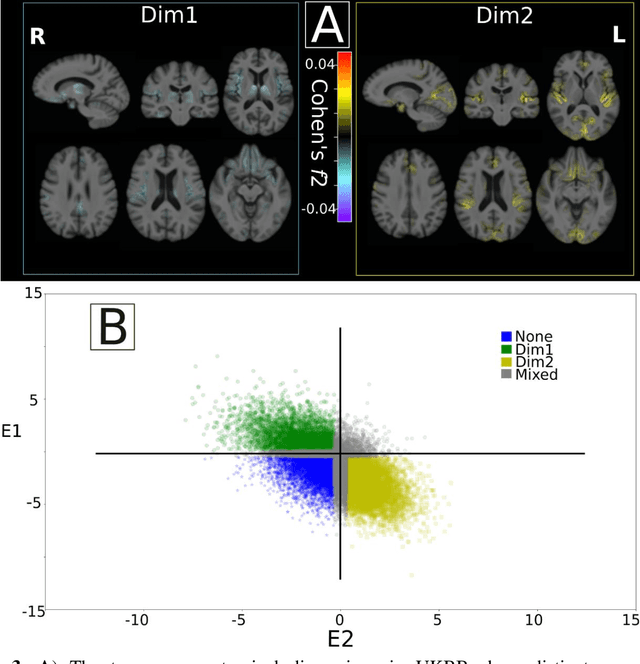
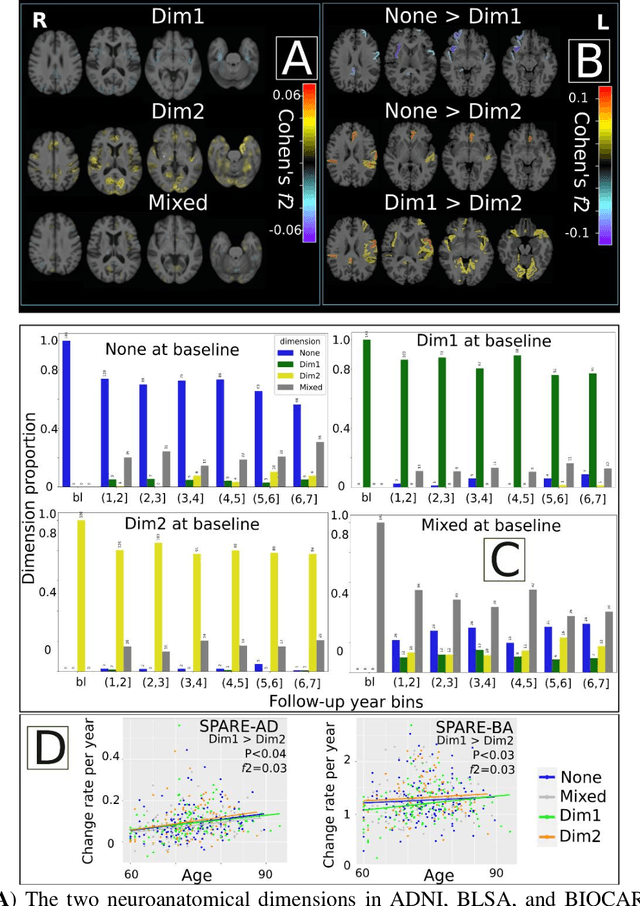
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge