Junhao Wen
for the Alzheimers Disease Neuroimaging Initiative
Gated Rotary-Enhanced Linear Attention for Long-term Sequential Recommendation
Jun 16, 2025Abstract:In Sequential Recommendation Systems (SRSs), Transformer models show remarkable performance but face computation cost challenges when modeling long-term user behavior sequences due to the quadratic complexity of the dot-product attention mechanism. By approximating the dot-product attention, linear attention provides an efficient option with linear complexity. However, existing linear attention methods face two limitations: 1) they often use learnable position encodings, which incur extra computational costs in long-term sequence scenarios, and 2) they may not consider the user's fine-grained local preferences and confuse these with the actual change of long-term interests. To remedy these drawbacks, we propose a long-term sequential Recommendation model with Gated Rotary Enhanced Linear Attention (RecGRELA). Specifically, we first propose a Rotary-Enhanced Linear Attention (RELA) module to model long-range dependency within the user's historical information using rotary position encodings. We then introduce a local short operation to incorporate local preferences and demonstrate the theoretical insight. We further introduce a SiLU-based Gated mechanism for RELA (GRELA) to help the model determine whether a user's behavior indicates local interest or a genuine shift in long-term preferences. Experimental results on four public datasets demonstrate that our RecGRELA achieves state-of-the-art performance compared to existing SRSs while maintaining low memory overhead.
Distance-aware Self-adaptive Graph Convolution for Fine-grained Hierarchical Recommendation
May 14, 2025Abstract:Graph Convolutional Networks (GCNs) are widely used to improve recommendation accuracy and performance by effectively learning the representations of user and item nodes. However, two major challenges remain: (1) the lack of further optimization in the graph representation structure and (2) insufficient attention given to the varying contributions of different convolutional layers.This paper proposes SAGCN, a distance-based adaptive hierarchical aggregation method that refines the aggregation process through differentiated representation metrics. SAGCN introduces a detailed approach to multilayer information aggregation and representation space optimization, enabling the model to learn hierarchical embedding weights based on the distance between hierarchical representations. This innovation allows for more precise cross-layer information aggregation, improves the model's ability to capture hierarchical embeddings, and optimizes the representation space structure. Additionally, the objective loss function is refined to better align with recommendation tasks.Extensive experiments conducted on four real-world datasets demonstrate significant improvements, including over a 5% increase on Yelp and a 5.58% increase in Recall@10 on the ML_1M dataset.
Breaking the Clusters: Uniformity-Optimization for Text-Based Sequential Recommendation
Feb 19, 2025



Abstract:Traditional sequential recommendation (SR) methods heavily rely on explicit item IDs to capture user preferences over time. This reliance introduces critical limitations in cold-start scenarios and domain transfer tasks, where unseen items and new contexts often lack established ID mappings. To overcome these limitations, recent studies have shifted towards leveraging text-only information for recommendation, thereby improving model generalization and adaptability across domains. Although promising, text-based SR faces unique difficulties: items' text descriptions often share semantic similarities that lead to clustered item representations, compromising their uniformity, a property essential for promoting diversity and enhancing generalization in recommendation systems. In this paper, we explore a novel framework to improve the uniformity of item representations in text-based SR. Our analysis reveals that items within a sequence exhibit marked semantic similarity, meaning they are closer in representation than items overall, and that this effect is more pronounced for less popular items, which form tighter clusters compared to their more popular counterparts. Based on these findings, we propose UniT, a framework that employs three pairwise item sampling strategies: Unified General Sampling Strategy, Sequence-Driven Sampling Strategy, and Popularity-Driven Sampling Strategy. Each strategy applies varying degrees of repulsion to selectively adjust the distances between item pairs, thereby refining representation uniformity while considering both sequence context and item popularity. Extensive experiments on multiple real-world datasets demonstrate that our proposed approach outperforms state-of-the-art models, validating the effectiveness of UniT in enhancing both representation uniformity and recommendation accuracy.The source code is available at https://github.com/ccwwhhh/Model-Rec.
LLM4SBR: A Lightweight and Effective Framework for Integrating Large Language Models in Session-based Recommendation
Feb 21, 2024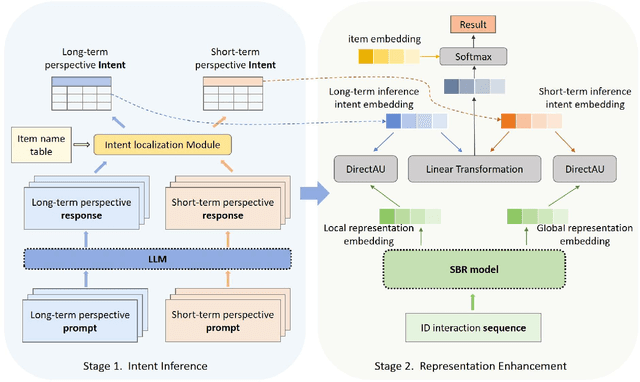

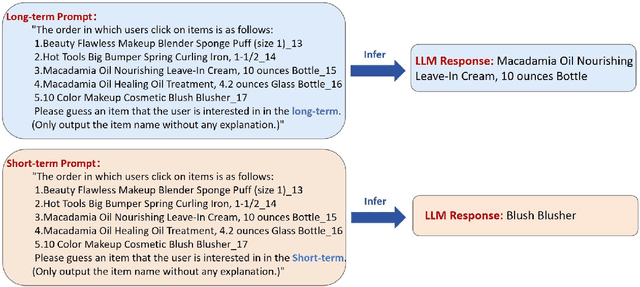
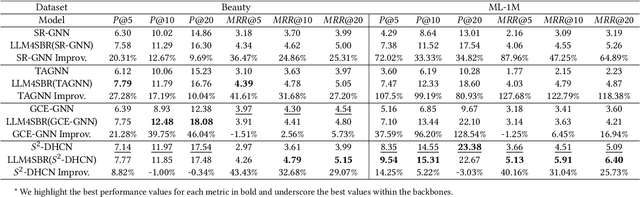
Abstract:Traditional session-based recommendation (SBR) utilizes session behavior sequences from anonymous users for recommendation. Although this strategy is highly efficient, it sacrifices the inherent semantic information of the items, making it difficult for the model to understand the true intent of the session and resulting in a lack of interpretability in the recommended results. Recently, large language models (LLMs) have flourished across various domains, offering a glimpse of hope in addressing the aforementioned challenges. Inspired by the impact of LLMs, research exploring the integration of LLMs with the Recommender system (RS) has surged like mushrooms after rain. However, constrained by high time and space costs, as well as the brief and anonymous nature of session data, the first LLM recommendation framework suitable for industrial deployment has yet to emerge in the field of SBR. To address the aforementioned challenges, we have proposed the LLM Integration Framework for SBR (LLM4SBR). Serving as a lightweight and plug-and-play framework, LLM4SBR adopts a two-step strategy. Firstly, we transform session data into a bimodal form of text and behavior. In the first step, leveraging the inferential capabilities of LLMs, we conduct inference on session text data from different perspectives and design the component for auxiliary enhancement. In the second step, the SBR model is trained on behavior data, aligning and averaging two modal session representations from different perspectives. Finally, we fuse session representations from different perspectives and modalities as the ultimate session representation for recommendation. We conducted experiments on two real-world datasets, and the results demonstrate that LLM4SBR significantly improves the performance of traditional SBR models and is highly lightweight and efficient, making it suitable for industrial deployment.
Dimensional Neuroimaging Endophenotypes: Neurobiological Representations of Disease Heterogeneity Through Machine Learning
Jan 17, 2024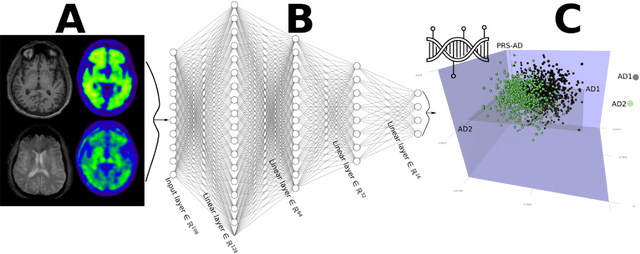
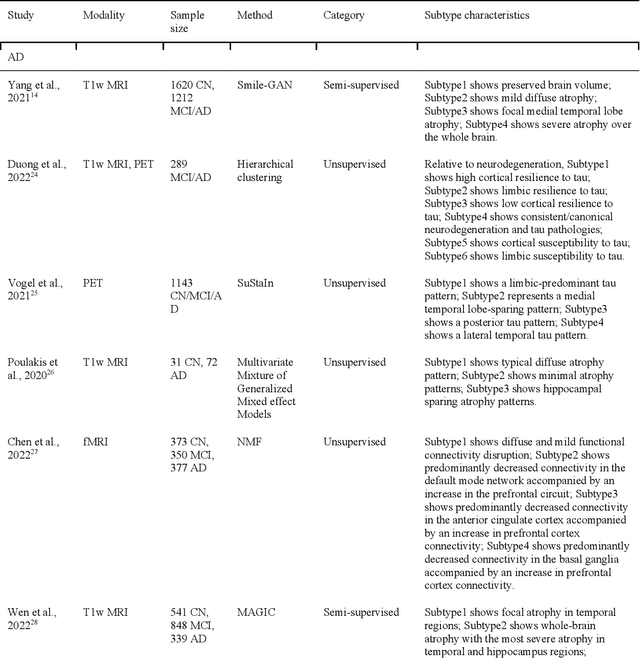
Abstract:Machine learning has been increasingly used to obtain individualized neuroimaging signatures for disease diagnosis, prognosis, and response to treatment in neuropsychiatric and neurodegenerative disorders. Therefore, it has contributed to a better understanding of disease heterogeneity by identifying disease subtypes that present significant differences in various brain phenotypic measures. In this review, we first present a systematic literature overview of studies using machine learning and multimodal MRI to unravel disease heterogeneity in various neuropsychiatric and neurodegenerative disorders, including Alzheimer disease, schizophrenia, major depressive disorder, autism spectrum disorder, multiple sclerosis, as well as their potential in transdiagnostic settings. Subsequently, we summarize relevant machine learning methodologies and discuss an emerging paradigm which we call dimensional neuroimaging endophenotype (DNE). DNE dissects the neurobiological heterogeneity of neuropsychiatric and neurodegenerative disorders into a low dimensional yet informative, quantitative brain phenotypic representation, serving as a robust intermediate phenotype (i.e., endophenotype) largely reflecting underlying genetics and etiology. Finally, we discuss the potential clinical implications of the current findings and envision future research avenues.
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023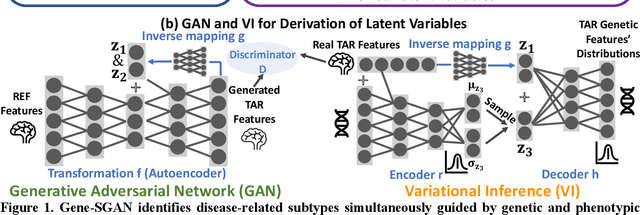
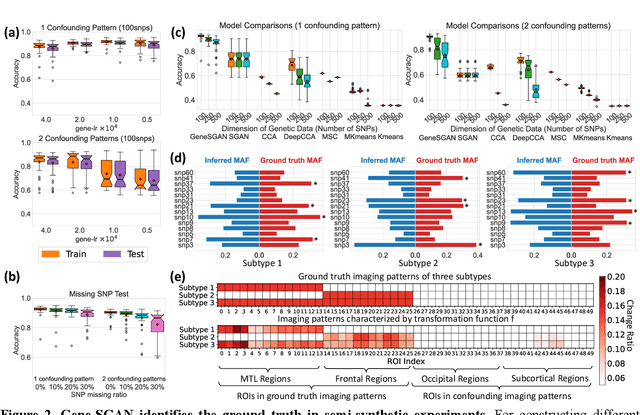
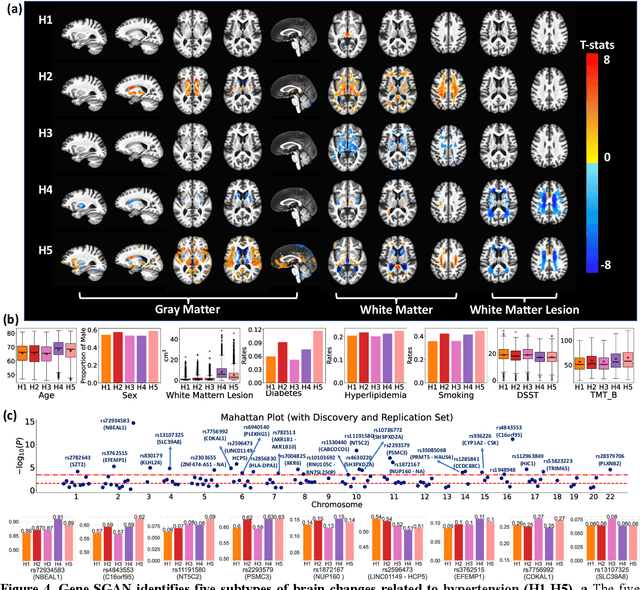
Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Applications of Generative Adversarial Networks in Neuroimaging and Clinical Neuroscience
Jun 14, 2022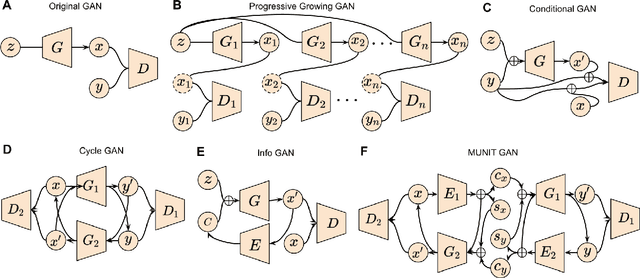
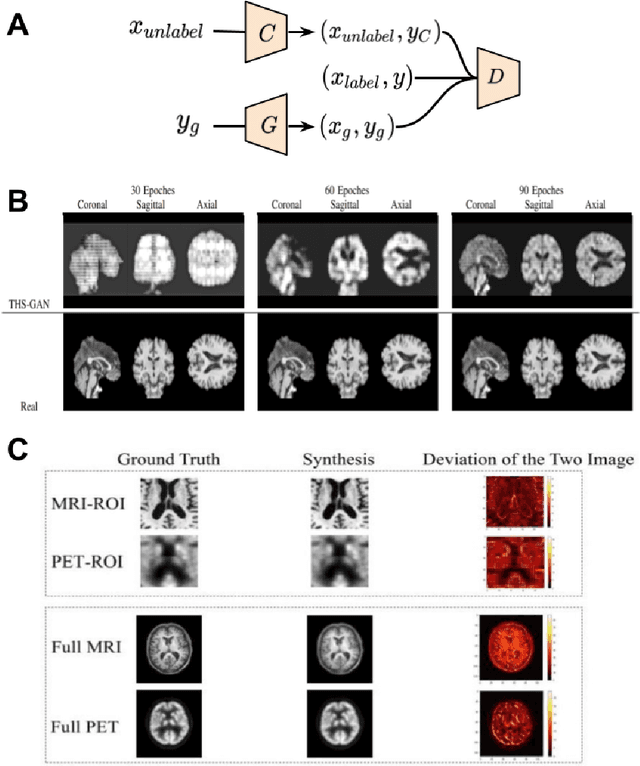
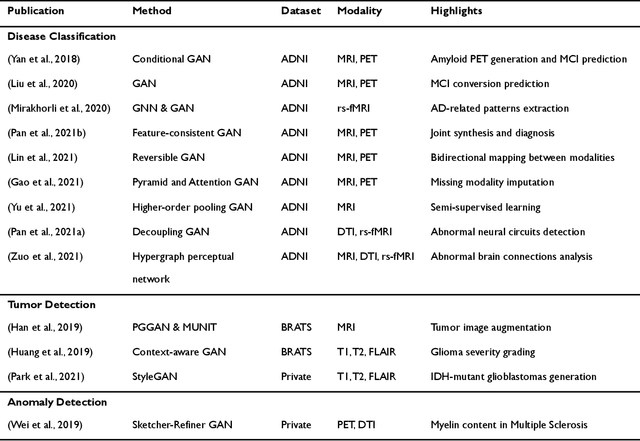
Abstract:Generative adversarial networks (GANs) are one powerful type of deep learning models that have been successfully utilized in numerous fields. They belong to a broader family called generative methods, which generate new data with a probabilistic model by learning sample distribution from real examples. In the clinical context, GANs have shown enhanced capabilities in capturing spatially complex, nonlinear, and potentially subtle disease effects compared to traditional generative methods. This review appraises the existing literature on the applications of GANs in imaging studies of various neurological conditions, including Alzheimer's disease, brain tumors, brain aging, and multiple sclerosis. We provide an intuitive explanation of various GAN methods for each application and further discuss the main challenges, open questions, and promising future directions of leveraging GANs in neuroimaging. We aim to bridge the gap between advanced deep learning methods and neurology research by highlighting how GANs can be leveraged to support clinical decision making and contribute to a better understanding of the structural and functional patterns of brain diseases.
Surreal-GAN:Semi-Supervised Representation Learning via GAN for uncovering heterogeneous disease-related imaging patterns
May 09, 2022



Abstract:A plethora of machine learning methods have been applied to imaging data, enabling the construction of clinically relevant imaging signatures of neurological and neuropsychiatric diseases. Oftentimes, such methods don't explicitly model the heterogeneity of disease effects, or approach it via nonlinear models that are not interpretable. Moreover, unsupervised methods may parse heterogeneity that is driven by nuisance confounding factors that affect brain structure or function, rather than heterogeneity relevant to a pathology of interest. On the other hand, semi-supervised clustering methods seek to derive a dichotomous subtype membership, ignoring the truth that disease heterogeneity spatially and temporally extends along a continuum. To address the aforementioned limitations, herein, we propose a novel method, termed Surreal-GAN (Semi-SUpeRvised ReprEsentAtion Learning via GAN). Using cross-sectional imaging data, Surreal-GAN dissects underlying disease-related heterogeneity under the principle of semi-supervised clustering (cluster mappings from normal control to patient), proposes a continuously dimensional representation, and infers the disease severity of patients at individual level along each dimension. The model first learns a transformation function from normal control (CN) domain to the patient (PT) domain with latent variables controlling transformation directions. An inverse mapping function together with regularization on function continuity, pattern orthogonality and monotonicity was also imposed to make sure that the transformation function captures necessarily meaningful imaging patterns with clinical significance. We first validated the model through extensive semi-synthetic experiments, and then demonstrate its potential in capturing biologically plausible imaging patterns in Alzheimer's disease (AD).
Subtyping brain diseases from imaging data
Feb 16, 2022

Abstract:The imaging community has increasingly adopted machine learning (ML) methods to provide individualized imaging signatures related to disease diagnosis, prognosis, and response to treatment. Clinical neuroscience and cancer imaging have been two areas in which ML has offered particular promise. However, many neurologic and neuropsychiatric diseases, as well as cancer, are often heterogeneous in terms of their clinical manifestations, neuroanatomical patterns or genetic underpinnings. Therefore, in such cases, seeking a single disease signature might be ineffectual in delivering individualized precision diagnostics. The current chapter focuses on ML methods, especially semi-supervised clustering, that seek disease subtypes using imaging data. Work from Alzheimer Disease and its prodromal stages, psychosis, depression, autism, and brain cancer are discussed. Our goal is to provide the readers with a broad overview in terms of methodology and clinical applications.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021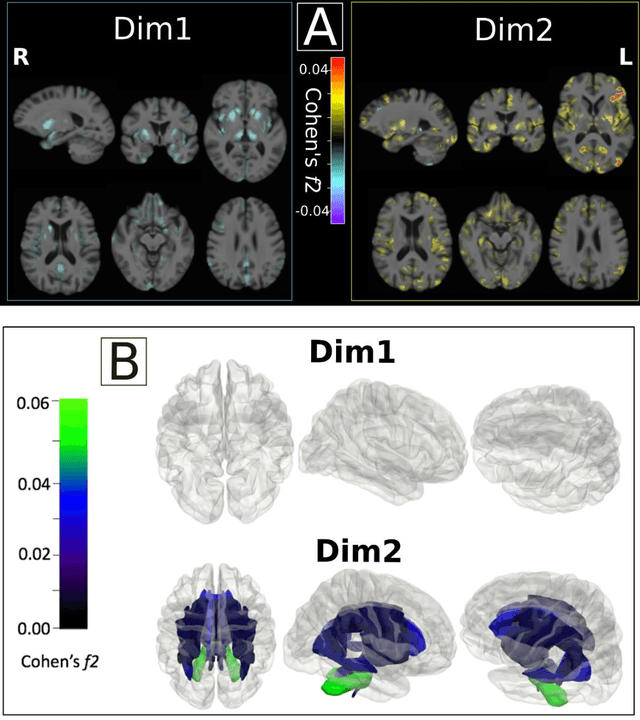
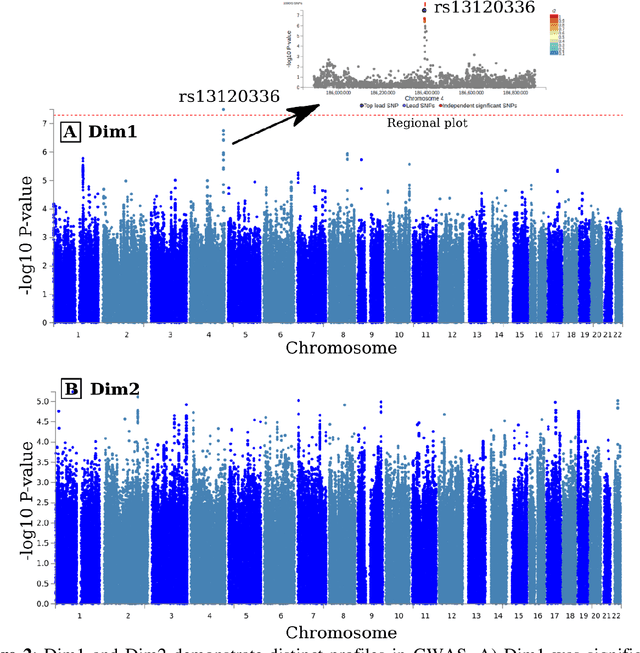
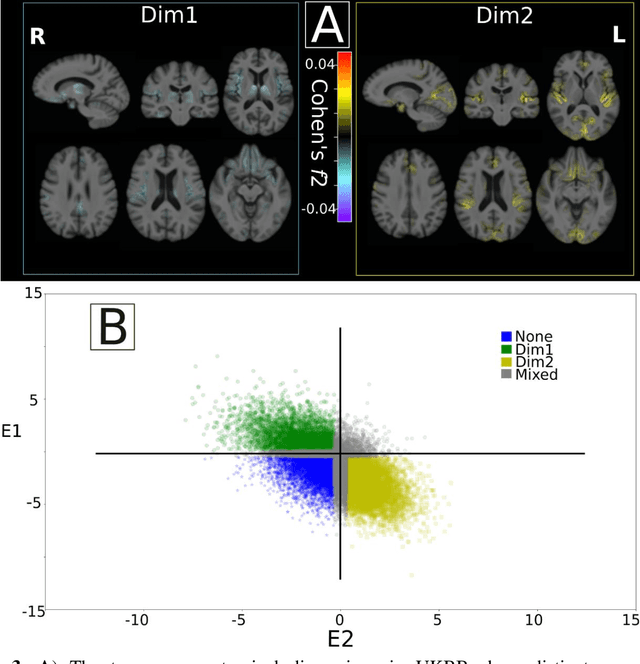
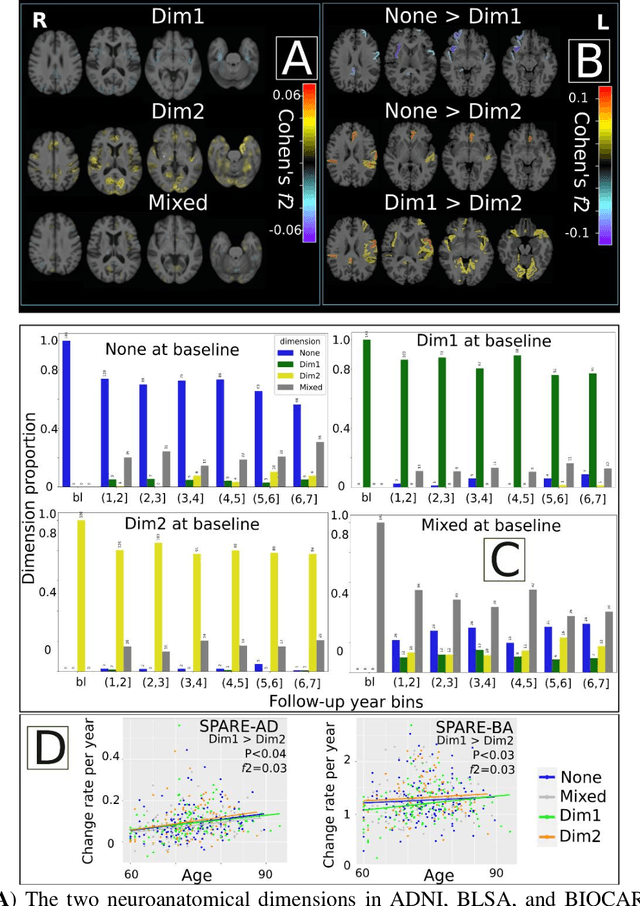
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge