Rongguang Wang
Fidelity-Imposed Displacement Editing for the Learn2Reg 2024 SHG-BF Challenge
Oct 28, 2024



Abstract:Co-examination of second-harmonic generation (SHG) and bright-field (BF) microscopy enables the differentiation of tissue components and collagen fibers, aiding the analysis of human breast and pancreatic cancer tissues. However, large discrepancies between SHG and BF images pose challenges for current learning-based registration models in aligning SHG to BF. In this paper, we propose a novel multi-modal registration framework that employs fidelity-imposed displacement editing to address these challenges. The framework integrates batch-wise contrastive learning, feature-based pre-alignment, and instance-level optimization. Experimental results from the Learn2Reg COMULISglobe SHG-BF Challenge validate the effectiveness of our method, securing the 1st place on the online leaderboard.
NeuroSynth: MRI-Derived Neuroanatomical Generative Models and Associated Dataset of 18,000 Samples
Jul 17, 2024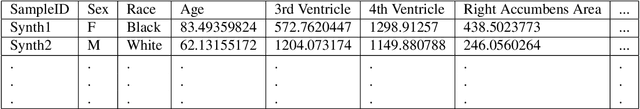
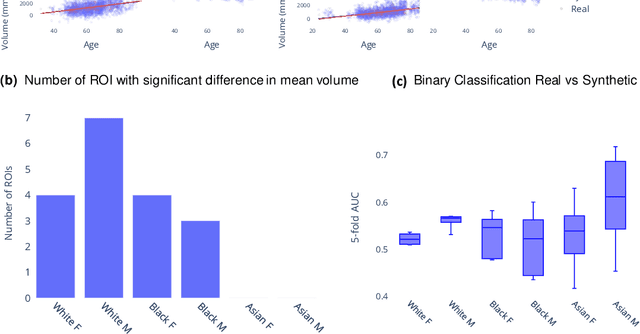
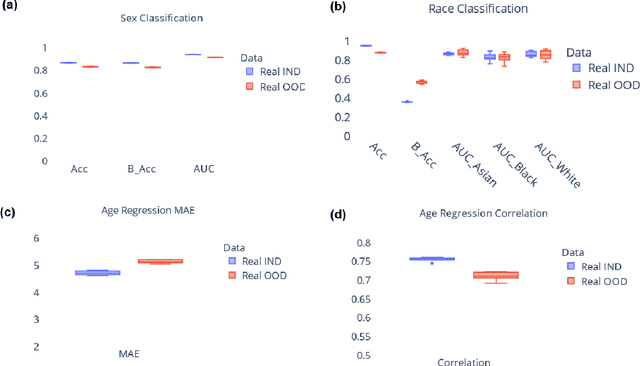
Abstract:Availability of large and diverse medical datasets is often challenged by privacy and data sharing restrictions. For successful application of machine learning techniques for disease diagnosis, prognosis, and precision medicine, large amounts of data are necessary for model building and optimization. To help overcome such limitations in the context of brain MRI, we present NeuroSynth: a collection of generative models of normative regional volumetric features derived from structural brain imaging. NeuroSynth models are trained on real brain imaging regional volumetric measures from the iSTAGING consortium, which encompasses over 40,000 MRI scans across 13 studies, incorporating covariates such as age, sex, and race. Leveraging NeuroSynth, we produce and offer 18,000 synthetic samples spanning the adult lifespan (ages 22-90 years), alongside the model's capability to generate unlimited data. Experimental results indicate that samples generated from NeuroSynth agree with the distributions obtained from real data. Most importantly, the generated normative data significantly enhance the accuracy of downstream machine learning models on tasks such as disease classification. Data and models are available at: https://huggingface.co/spaces/rongguangw/neuro-synth.
MemWarp: Discontinuity-Preserving Cardiac Registration with Memorized Anatomical Filters
Jul 10, 2024Abstract:Many existing learning-based deformable image registration methods impose constraints on deformation fields to ensure they are globally smooth and continuous. However, this assumption does not hold in cardiac image registration, where different anatomical regions exhibit asymmetric motions during respiration and movements due to sliding organs within the chest. Consequently, such global constraints fail to accommodate local discontinuities across organ boundaries, potentially resulting in erroneous and unrealistic displacement fields. In this paper, we address this issue with MemWarp, a learning framework that leverages a memory network to store prototypical information tailored to different anatomical regions. MemWarp is different from earlier approaches in two main aspects: firstly, by decoupling feature extraction from similarity matching in moving and fixed images, it facilitates more effective utilization of feature maps; secondly, despite its capability to preserve discontinuities, it eliminates the need for segmentation masks during model inference. In experiments on a publicly available cardiac dataset, our method achieves considerable improvements in registration accuracy and producing realistic deformations, outperforming state-of-the-art methods with a remarkable 7.1\% Dice score improvement over the runner-up semi-supervised method. Source code will be available at https://github.com/tinymilky/Mem-Warp.
Slicer Networks
Jan 18, 2024Abstract:In medical imaging, scans often reveal objects with varied contrasts but consistent internal intensities or textures. This characteristic enables the use of low-frequency approximations for tasks such as segmentation and deformation field estimation. Yet, integrating this concept into neural network architectures for medical image analysis remains underexplored. In this paper, we propose the Slicer Network, a novel architecture designed to leverage these traits. Comprising an encoder utilizing models like vision transformers for feature extraction and a slicer employing a learnable bilateral grid, the Slicer Network strategically refines and upsamples feature maps via a splatting-blurring-slicing process. This introduces an edge-preserving low-frequency approximation for the network outcome, effectively enlarging the effective receptive field. The enhancement not only reduces computational complexity but also boosts overall performance. Experiments across different medical imaging applications, including unsupervised and keypoints-based image registration and lesion segmentation, have verified the Slicer Network's improved accuracy and efficiency.
Dimensional Neuroimaging Endophenotypes: Neurobiological Representations of Disease Heterogeneity Through Machine Learning
Jan 17, 2024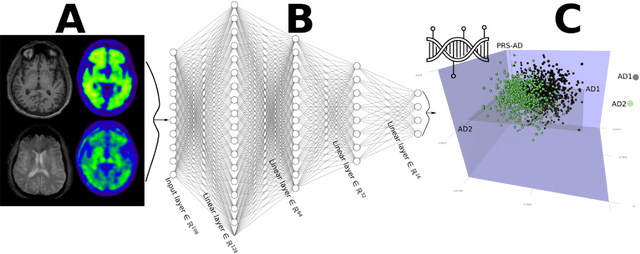
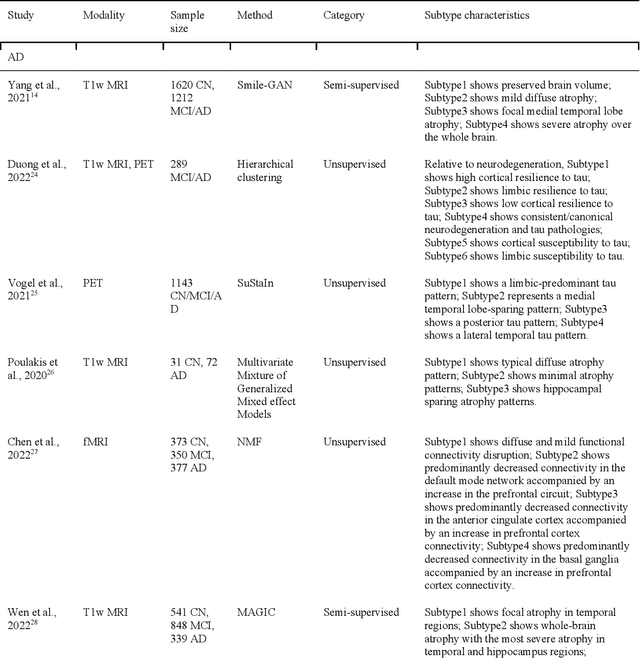
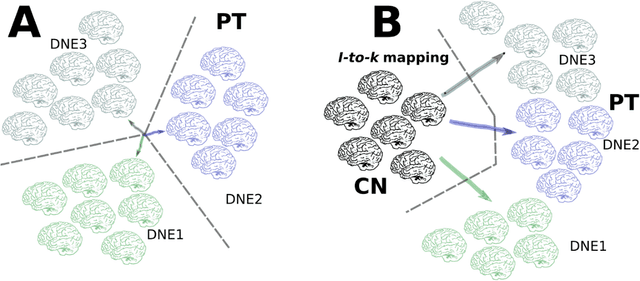
Abstract:Machine learning has been increasingly used to obtain individualized neuroimaging signatures for disease diagnosis, prognosis, and response to treatment in neuropsychiatric and neurodegenerative disorders. Therefore, it has contributed to a better understanding of disease heterogeneity by identifying disease subtypes that present significant differences in various brain phenotypic measures. In this review, we first present a systematic literature overview of studies using machine learning and multimodal MRI to unravel disease heterogeneity in various neuropsychiatric and neurodegenerative disorders, including Alzheimer disease, schizophrenia, major depressive disorder, autism spectrum disorder, multiple sclerosis, as well as their potential in transdiagnostic settings. Subsequently, we summarize relevant machine learning methodologies and discuss an emerging paradigm which we call dimensional neuroimaging endophenotype (DNE). DNE dissects the neurobiological heterogeneity of neuropsychiatric and neurodegenerative disorders into a low dimensional yet informative, quantitative brain phenotypic representation, serving as a robust intermediate phenotype (i.e., endophenotype) largely reflecting underlying genetics and etiology. Finally, we discuss the potential clinical implications of the current findings and envision future research avenues.
Spatially Covariant Image Registration with Text Prompts
Nov 27, 2023



Abstract:Medical images are often characterized by their structured anatomical representations and spatially inhomogeneous contrasts. Leveraging anatomical priors in neural networks can greatly enhance their utility in resource-constrained clinical settings. Prior research has harnessed such information for image segmentation, yet progress in deformable image registration has been modest. Our work introduces textSCF, a novel method that integrates spatially covariant filters and textual anatomical prompts encoded by visual-language models, to fill this gap. This approach optimizes an implicit function that correlates text embeddings of anatomical regions to filter weights, relaxing the typical translation-invariance constraint of convolutional operations. TextSCF not only boosts computational efficiency but can also retain or improve registration accuracy. By capturing the contextual interplay between anatomical regions, it offers impressive inter-regional transferability and the ability to preserve structural discontinuities during registration. TextSCF's performance has been rigorously tested on inter-subject brain MRI and abdominal CT registration tasks, outperforming existing state-of-the-art models in the MICCAI Learn2Reg 2021 challenge and leading the leaderboard. In abdominal registrations, textSCF's larger model variant improved the Dice score by 11.3% over the second-best model, while its smaller variant maintained similar accuracy but with an 89.13% reduction in network parameters and a 98.34\% decrease in computational operations.
Adapting Machine Learning Diagnostic Models to New Populations Using a Small Amount of Data: Results from Clinical Neuroscience
Aug 06, 2023



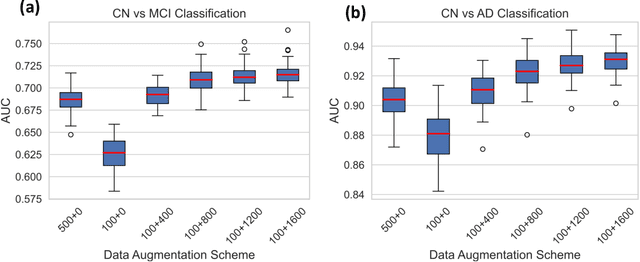
Abstract:Machine learning (ML) has shown great promise for revolutionizing a number of areas, including healthcare. However, it is also facing a reproducibility crisis, especially in medicine. ML models that are carefully constructed from and evaluated on a training set might not generalize well on data from different patient populations or acquisition instrument settings and protocols. We tackle this problem in the context of neuroimaging of Alzheimer's disease (AD), schizophrenia (SZ) and brain aging. We develop a weighted empirical risk minimization approach that optimally combines data from a source group, e.g., subjects are stratified by attributes such as sex, age group, race and clinical cohort to make predictions on a target group, e.g., other sex, age group, etc. using a small fraction (10%) of data from the target group. We apply this method to multi-source data of 15,363 individuals from 20 neuroimaging studies to build ML models for diagnosis of AD and SZ, and estimation of brain age. We found that this approach achieves substantially better accuracy than existing domain adaptation techniques: it obtains area under curve greater than 0.95 for AD classification, area under curve greater than 0.7 for SZ classification and mean absolute error less than 5 years for brain age prediction on all target groups, achieving robustness to variations of scanners, protocols, and demographic or clinical characteristics. In some cases, it is even better than training on all data from the target group, because it leverages the diversity and size of a larger training set. We also demonstrate the utility of our models for prognostic tasks such as predicting disease progression in individuals with mild cognitive impairment. Critically, our brain age prediction models lead to new clinical insights regarding correlations with neurophysiological tests.
DAGrid: Directed Accumulator Grid
Jun 05, 2023Abstract:Recent research highlights that the Directed Accumulator (DA), through its parametrization of geometric priors into neural networks, has notably improved the performance of medical image recognition, particularly with small and imbalanced datasets. However, DA's potential in pixel-wise dense predictions is unexplored. To bridge this gap, we present the Directed Accumulator Grid (DAGrid), which allows geometric-preserving filtering in neural networks, thus broadening the scope of DA's applications to include pixel-level dense prediction tasks. DAGrid utilizes homogeneous data types in conjunction with designed sampling grids to construct geometrically transformed representations, retaining intricate geometric information and promoting long-range information propagation within the neural networks. Contrary to its symmetric counterpart, grid sampling, which might lose information in the sampling process, DAGrid aggregates all pixels, ensuring a comprehensive representation in the transformed space. The parallelization of DAGrid on modern GPUs is facilitated using CUDA programming, and also back propagation is enabled for deep neural network training. Empirical results show DAGrid-enhanced neural networks excel in supervised skin lesion segmentation and unsupervised cardiac image registration. Specifically, the network incorporating DAGrid has realized a 70.8% reduction in network parameter size and a 96.8% decrease in FLOPs, while concurrently improving the Dice score for skin lesion segmentation by 1.0% compared to state-of-the-art transformers. Furthermore, it has achieved improvements of 4.4% and 8.2% in the average Dice score and Dice score of the left ventricular mass, respectively, indicating an increase in registration accuracy for cardiac images. The source code is available at https://github.com/tinymilky/DeDA.
DeDA: Deep Directed Accumulator
Mar 15, 2023Abstract:Chronic active multiple sclerosis lesions, also termed as rim+ lesions, can be characterized by a hyperintense rim at the edge of the lesion on quantitative susceptibility maps. These rim+ lesions exhibit a geometrically simple structure, where gradients at the lesion edge are radially oriented and a greater magnitude of gradients is observed in contrast to rim- (non rim+) lesions. However, recent studies have shown that the identification performance of such lesions remains unsatisfied due to the limited amount of data and high class imbalance. In this paper, we propose a simple yet effective image processing operation, deep directed accumulator (DeDA), that provides a new perspective for injecting domain-specific inductive biases (priors) into neural networks for rim+ lesion identification. Given a feature map and a set of sampling grids, DeDA creates and quantizes an accumulator space into finite intervals, and accumulates feature values accordingly. This DeDA operation is a generalized discrete Radon transform and can also be regarded as a symmetric operation to the grid sampling within the forward-backward neural network framework, the process of which is order-agnostic, and can be efficiently implemented with the native CUDA programming. Experimental results on a dataset with 177 rim+ and 3986 rim- lesions show that 10.1% of improvement in a partial (false positive rate<0.1) area under the receiver operating characteristic curve (pROC AUC) and 10.2% of improvement in an area under the precision recall curve (PR AUC) can be achieved respectively comparing to other state-of-the-art methods. The source code is available online at https://github.com/tinymilky/DeDA
Spatially Covariant Lesion Segmentation
Jan 19, 2023



Abstract:Compared to natural images, medical images usually show stronger visual patterns and therefore this adds flexibility and elasticity to resource-limited clinical applications by injecting proper priors into neural networks. In this paper, we propose spatially covariant pixel-aligned classifier (SCP) to improve the computational efficiency and meantime maintain or increase accuracy for lesion segmentation. SCP relaxes the spatial invariance constraint imposed by convolutional operations and optimizes an underlying implicit function that maps image coordinates to network weights, the parameters of which are obtained along with the backbone network training and later used for generating network weights to capture spatially covariant contextual information. We demonstrate the effectiveness and efficiency of the proposed SCP using two lesion segmentation tasks from different imaging modalities: white matter hyperintensity segmentation in magnetic resonance imaging and liver tumor segmentation in contrast-enhanced abdominal computerized tomography. The network using SCP has achieved 23.8%, 64.9% and 74.7% reduction in GPU memory usage, FLOPs, and network size with similar or better accuracy for lesion segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge