Jiazheng Wang
Unsupervised MRI-US Multimodal Image Registration with Multilevel Correlation Pyramidal Optimization
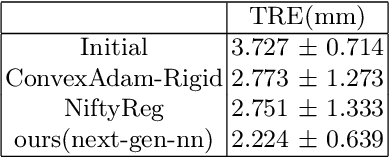
Feb 06, 2026Abstract:Surgical navigation based on multimodal image registration has played a significant role in providing intraoperative guidance to surgeons by showing the relative position of the target area to critical anatomical structures during surgery. However, due to the differences between multimodal images and intraoperative image deformation caused by tissue displacement and removal during the surgery, effective registration of preoperative and intraoperative multimodal images faces significant challenges. To address the multimodal image registration challenges in Learn2Reg 2025, an unsupervised multimodal medical image registration method based on multilevel correlation pyramidal optimization (MCPO) is designed to solve these problems. First, the features of each modality are extracted based on the modality independent neighborhood descriptor, and the multimodal images is mapped to the feature space. Second, a multilevel pyramidal fusion optimization mechanism is designed to achieve global optimization and local detail complementation of the displacement field through dense correlation analysis and weight-balanced coupled convex optimization for input features at different scales. Our method focuses on the ReMIND2Reg task in Learn2Reg 2025. Based on the results, our method achieved the first place in the validation phase and test phase of ReMIND2Reg. The MCPO is also validated on the Resect dataset, achieving an average TRE of 1.798 mm. This demonstrates the broad applicability of our method in preoperative-to-intraoperative image registration. The code is avaliable at https://github.com/wjiazheng/MCPO.
GeoLanG: Geometry-Aware Language-Guided Grasping with Unified RGB-D Multimodal Learning
Feb 04, 2026Abstract:Language-guided grasping has emerged as a promising paradigm for enabling robots to identify and manipulate target objects through natural language instructions, yet it remains highly challenging in cluttered or occluded scenes. Existing methods often rely on multi-stage pipelines that separate object perception and grasping, which leads to limited cross-modal fusion, redundant computation, and poor generalization in cluttered, occluded, or low-texture scenes. To address these limitations, we propose GeoLanG, an end-to-end multi-task framework built upon the CLIP architecture that unifies visual and linguistic inputs into a shared representation space for robust semantic alignment and improved generalization. To enhance target discrimination under occlusion and low-texture conditions, we explore a more effective use of depth information through the Depth-guided Geometric Module (DGGM), which converts depth into explicit geometric priors and injects them into the attention mechanism without additional computational overhead. In addition, we propose Adaptive Dense Channel Integration, which adaptively balances the contributions of multi-layer features to produce more discriminative and generalizable visual representations. Extensive experiments on the OCID-VLG dataset, as well as in both simulation and real-world hardware, demonstrate that GeoLanG enables precise and robust language-guided grasping in complex, cluttered environments, paving the way toward more reliable multimodal robotic manipulation in real-world human-centric settings.
A Step to Decouple Optimization in 3DGS
Jan 26, 2026Abstract:3D Gaussian Splatting (3DGS) has emerged as a powerful technique for real-time novel view synthesis. As an explicit representation optimized through gradient propagation among primitives, optimization widely accepted in deep neural networks (DNNs) is actually adopted in 3DGS, such as synchronous weight updating and Adam with the adaptive gradient. However, considering the physical significance and specific design in 3DGS, there are two overlooked details in the optimization of 3DGS: (i) update step coupling, which induces optimizer state rescaling and costly attribute updates outside the viewpoints, and (ii) gradient coupling in the moment, which may lead to under- or over-effective regularization. Nevertheless, such a complex coupling is under-explored. After revisiting the optimization of 3DGS, we take a step to decouple it and recompose the process into: Sparse Adam, Re-State Regularization and Decoupled Attribute Regularization. Taking a large number of experiments under the 3DGS and 3DGS-MCMC frameworks, our work provides a deeper understanding of these components. Finally, based on the empirical analysis, we re-design the optimization and propose AdamW-GS by re-coupling the beneficial components, under which better optimization efficiency and representation effectiveness are achieved simultaneously.
Gaussian Primitive Optimized Deformable Retinal Image Registration
Aug 23, 2025



Abstract:Deformable retinal image registration is notoriously difficult due to large homogeneous regions and sparse but critical vascular features, which cause limited gradient signals in standard learning-based frameworks. In this paper, we introduce Gaussian Primitive Optimization (GPO), a novel iterative framework that performs structured message passing to overcome these challenges. After an initial coarse alignment, we extract keypoints at salient anatomical structures (e.g., major vessels) to serve as a minimal set of descriptor-based control nodes (DCN). Each node is modelled as a Gaussian primitive with trainable position, displacement, and radius, thus adapting its spatial influence to local deformation scales. A K-Nearest Neighbors (KNN) Gaussian interpolation then blends and propagates displacement signals from these information-rich nodes to construct a globally coherent displacement field; focusing interpolation on the top (K) neighbors reduces computational overhead while preserving local detail. By strategically anchoring nodes in high-gradient regions, GPO ensures robust gradient flow, mitigating vanishing gradient signal in textureless areas. The framework is optimized end-to-end via a multi-term loss that enforces both keypoint consistency and intensity alignment. Experiments on the FIRE dataset show that GPO reduces the target registration error from 6.2\,px to ~2.4\,px and increases the AUC at 25\,px from 0.770 to 0.938, substantially outperforming existing methods. The source code can be accessed via https://github.com/xintian-99/GPOreg.
ShorterBetter: Guiding Reasoning Models to Find Optimal Inference Length for Efficient Reasoning
Apr 30, 2025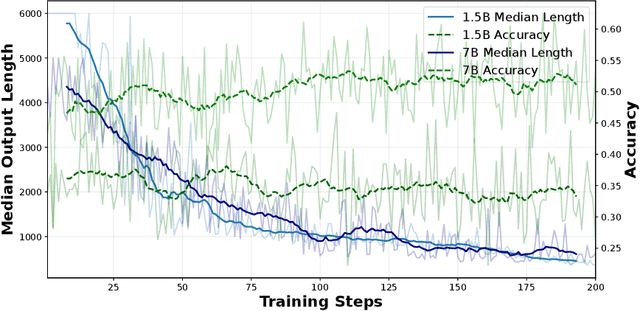
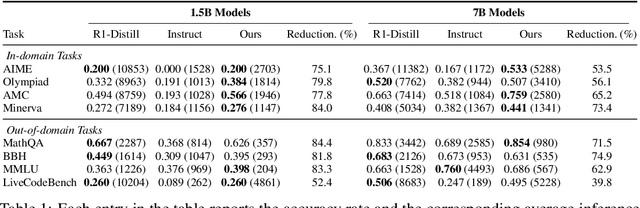
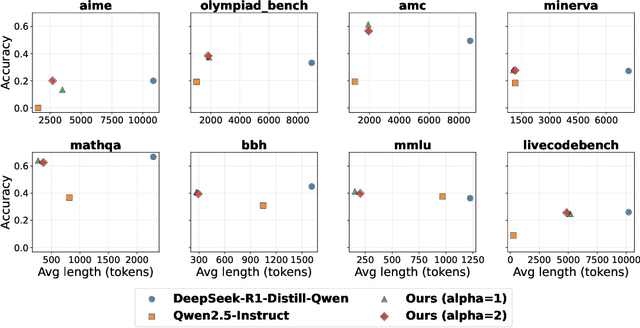
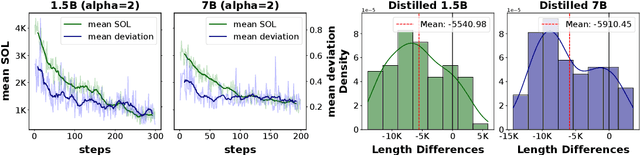
Abstract:Reasoning models such as OpenAI o3 and DeepSeek-R1 have demonstrated strong performance on reasoning-intensive tasks through extended Chain-of-Thought (CoT) prompting. While longer reasoning traces can facilitate a more thorough exploration of solution paths for complex problems, researchers have observed that these models often "overthink", leading to inefficient inference. In this paper, we introduce ShorterBetter, a simple yet effective reinforcement learning methed that enables reasoning language models to discover their own optimal CoT lengths without human intervention. By sampling multiple outputs per problem and defining the Sample Optimal Length (SOL) as the shortest correct response among all the outputs, our method dynamically guides the model toward optimal inference lengths. Applied to the DeepSeek-Distill-Qwen-1.5B model, ShorterBetter achieves up to an 80% reduction in output length on both in-domain and out-of-domain reasoning tasks while maintaining accuracy. Our analysis shows that overly long reasoning traces often reflect loss of reasoning direction, and thus suggests that the extended CoT produced by reasoning models is highly compressible.
Reproducibility Assessment of Magnetic Resonance Spectroscopy of Pregenual Anterior Cingulate Cortex across Sessions and Vendors via the Cloud Computing Platform CloudBrain-MRS
Mar 06, 2025



Abstract:Given the need to elucidate the mechanisms underlying illnesses and their treatment, as well as the lack of harmonization of acquisition and post-processing protocols among different magnetic resonance system vendors, this work is to determine if metabolite concentrations obtained from different sessions, machine models and even different vendors of 3 T scanners can be highly reproducible and be pooled for diagnostic analysis, which is very valuable for the research of rare diseases. Participants underwent magnetic resonance imaging (MRI) scanning once on two separate days within one week (one session per day, each session including two proton magnetic resonance spectroscopy (1H-MRS) scans with no more than a 5-minute interval between scans (no off-bed activity)) on each machine. were analyzed for reliability of within- and between- sessions using the coefficient of variation (CV) and intraclass correlation coefficient (ICC), and for reproducibility of across the machines using correlation coefficient. As for within- and between- session, all CV values for a group of all the first or second scans of a session, or for a session were almost below 20%, and most of the ICCs for metabolites range from moderate (0.4-0.59) to excellent (0.75-1), indicating high data reliability. When it comes to the reproducibility across the three scanners, all Pearson correlation coefficients across the three machines approached 1 with most around 0.9, and majority demonstrated statistical significance (P<0.01). Additionally, the intra-vendor reproducibility was greater than the inter-vendor ones.
EndoChat: Grounded Multimodal Large Language Model for Endoscopic Surgery
Jan 20, 2025
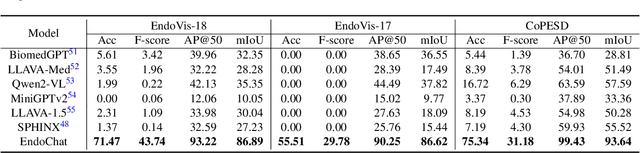
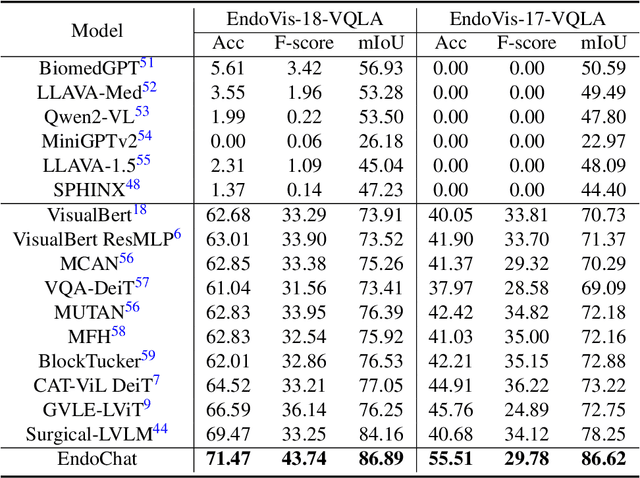
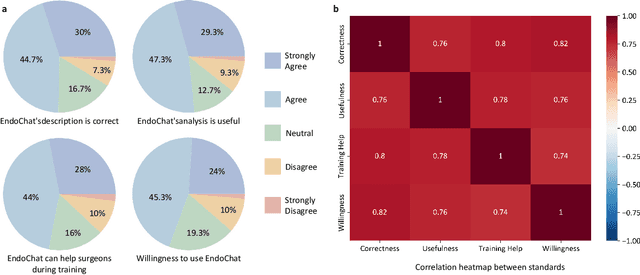
Abstract:Recently, Multimodal Large Language Models (MLLMs) have demonstrated their immense potential in computer-aided diagnosis and decision-making. In the context of robotic-assisted surgery, MLLMs can serve as effective tools for surgical training and guidance. However, there is still a lack of MLLMs specialized for surgical scene understanding in clinical applications. In this work, we introduce EndoChat to address various dialogue paradigms and subtasks in surgical scene understanding that surgeons encounter. To train our EndoChat, we construct the Surg-396K dataset through a novel pipeline that systematically extracts surgical information and generates structured annotations based on collected large-scale endoscopic surgery datasets. Furthermore, we introduce a multi-scale visual token interaction mechanism and a visual contrast-based reasoning mechanism to enhance the model's representation learning and reasoning capabilities. Our model achieves state-of-the-art performance across five dialogue paradigms and eight surgical scene understanding tasks. Additionally, we conduct evaluations with professional surgeons, most of whom provide positive feedback on collaborating with EndoChat. Overall, these results demonstrate that our EndoChat has great potential to significantly advance training and automation in robotic-assisted surgery.
Enhancing Transformers for Generalizable First-Order Logical Entailment
Jan 01, 2025



Abstract:Transformers, as a fundamental deep learning architecture, have demonstrated remarkable capabilities in reasoning. This paper investigates the generalizable first-order logical reasoning ability of transformers with their parameterized knowledge and explores ways to improve it. The first-order reasoning capability of transformers is assessed through their ability to perform first-order logical entailment, which is quantitatively measured by their performance in answering knowledge graph queries. We establish connections between (1) two types of distribution shifts studied in out-of-distribution generalization and (2) the unseen knowledge and query settings discussed in the task of knowledge graph query answering, enabling a characterization of fine-grained generalizability. Results on our comprehensive dataset show that transformers outperform previous methods specifically designed for this task and provide detailed empirical evidence on the impact of input query syntax, token embedding, and transformer architectures on the reasoning capability of transformers. Interestingly, our findings reveal a mismatch between positional encoding and other design choices in transformer architectures employed in prior practices. This discovery motivates us to propose a more sophisticated, logic-aware architecture, TEGA, to enhance the capability for generalizable first-order logical entailment in transformers.
Fidelity-Imposed Displacement Editing for the Learn2Reg 2024 SHG-BF Challenge
Oct 28, 2024



Abstract:Co-examination of second-harmonic generation (SHG) and bright-field (BF) microscopy enables the differentiation of tissue components and collagen fibers, aiding the analysis of human breast and pancreatic cancer tissues. However, large discrepancies between SHG and BF images pose challenges for current learning-based registration models in aligning SHG to BF. In this paper, we propose a novel multi-modal registration framework that employs fidelity-imposed displacement editing to address these challenges. The framework integrates batch-wise contrastive learning, feature-based pre-alignment, and instance-level optimization. Experimental results from the Learn2Reg COMULISglobe SHG-BF Challenge validate the effectiveness of our method, securing the 1st place on the online leaderboard.
Unsupervised Multimodal 3D Medical Image Registration with Multilevel Correlation Balanced Optimization
Sep 08, 2024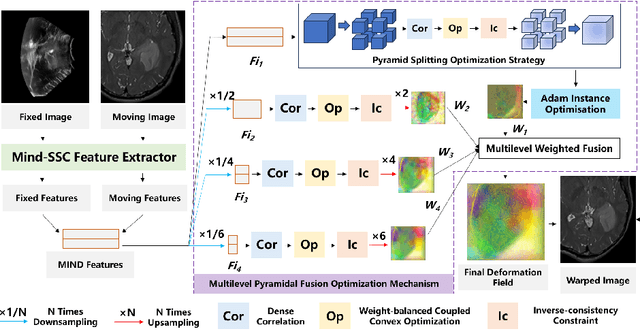
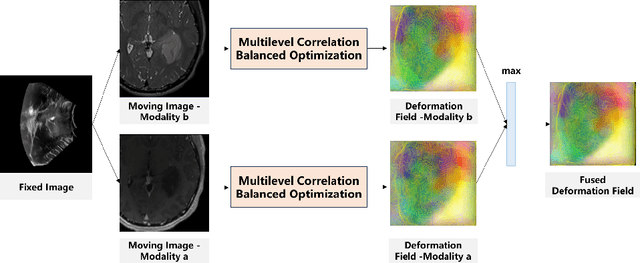

Abstract:Surgical navigation based on multimodal image registration has played a significant role in providing intraoperative guidance to surgeons by showing the relative position of the target area to critical anatomical structures during surgery. However, due to the differences between multimodal images and intraoperative image deformation caused by tissue displacement and removal during the surgery, effective registration of preoperative and intraoperative multimodal images faces significant challenges. To address the multimodal image registration challenges in Learn2Reg 2024, an unsupervised multimodal medical image registration method based on multilevel correlation balanced optimization (MCBO) is designed to solve these problems. First, the features of each modality are extracted based on the modality independent neighborhood descriptor, and the multimodal images is mapped to the feature space. Second, a multilevel pyramidal fusion optimization mechanism is designed to achieve global optimization and local detail complementation of the deformation field through dense correlation analysis and weight-balanced coupled convex optimization for input features at different scales. For preoperative medical images in different modalities, the alignment and stacking of valid information between different modalities is achieved by the maximum fusion between deformation fields. Our method focuses on the ReMIND2Reg task in Learn2Reg 2024, and to verify the generality of the method, we also tested it on the COMULIS3DCLEM task. Based on the results, our method achieved second place in the validation of both two tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge