Dicheng Chen
An artificially intelligent magnetic resonance spectroscopy quantification method: Comparison between QNet and LCModel on the cloud computing platform CloudBrain-MRS
Mar 06, 2025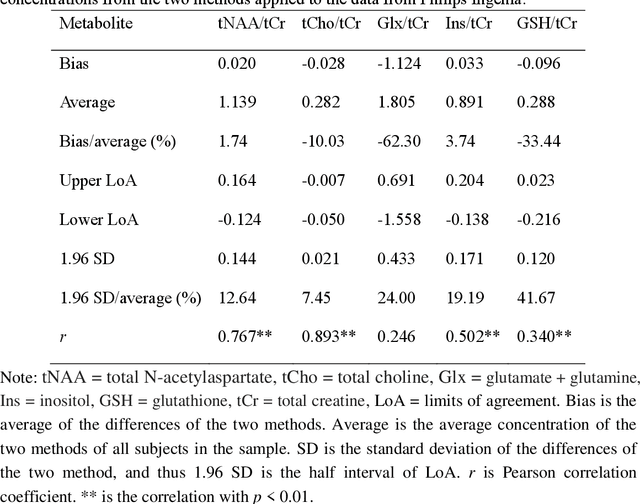
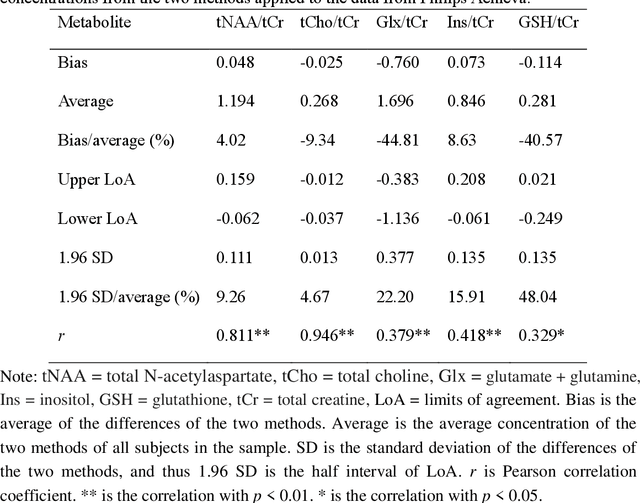
Abstract:Objctives: This work aimed to statistically compare the metabolite quantification of human brain magnetic resonance spectroscopy (MRS) between the deep learning method QNet and the classical method LCModel through an easy-to-use intelligent cloud computing platform CloudBrain-MRS. Materials and Methods: In this retrospective study, two 3 T MRI scanners Philips Ingenia and Achieva collected 61 and 46 in vivo 1H magnetic resonance (MR) spectra of healthy participants, respectively, from the brain region of pregenual anterior cingulate cortex from September to October 2021. The analyses of Bland-Altman, Pearson correlation and reasonability were performed to assess the degree of agreement, linear correlation and reasonability between the two quantification methods. Results: Fifteen healthy volunteers (12 females and 3 males, age range: 21-35 years, mean age/standard deviation = 27.4/3.9 years) were recruited. The analyses of Bland-Altman, Pearson correlation and reasonability showed high to good consistency and very strong to moderate correlation between the two methods for quantification of total N-acetylaspartate (tNAA), total choline (tCho), and inositol (Ins) (relative half interval of limits of agreement = 3.04%, 9.3%, and 18.5%, respectively; Pearson correlation coefficient r = 0.775, 0.927, and 0.469, respectively). In addition, quantification results of QNet are more likely to be closer to the previous reported average values than those of LCModel. Conclusion: There were high or good degrees of consistency between the quantification results of QNet and LCModel for tNAA, tCho, and Ins, and QNet generally has more reasonable quantification than LCModel.
CloudBrain-MRS: An Intelligent Cloud Computing Platform for in vivo Magnetic Resonance Spectroscopy Preprocessing, Quantification, and Analysis
Jun 19, 2023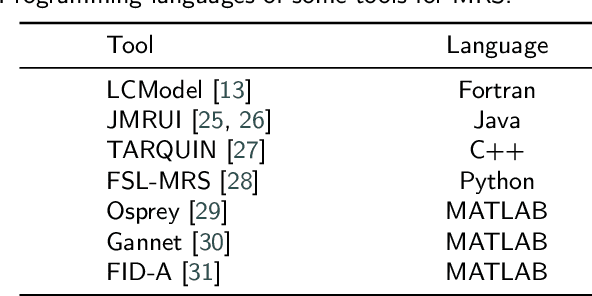
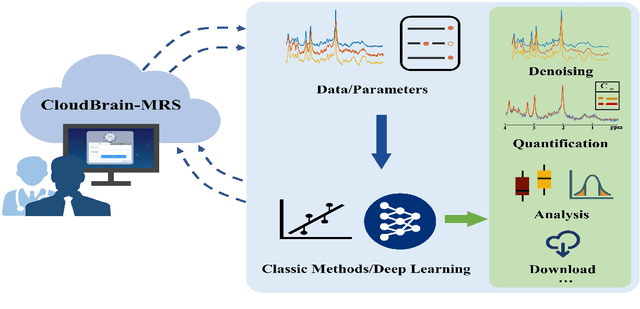
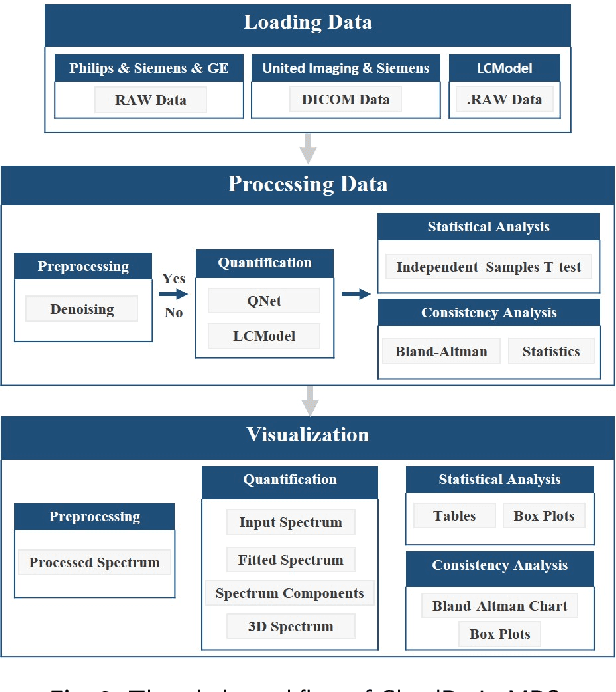
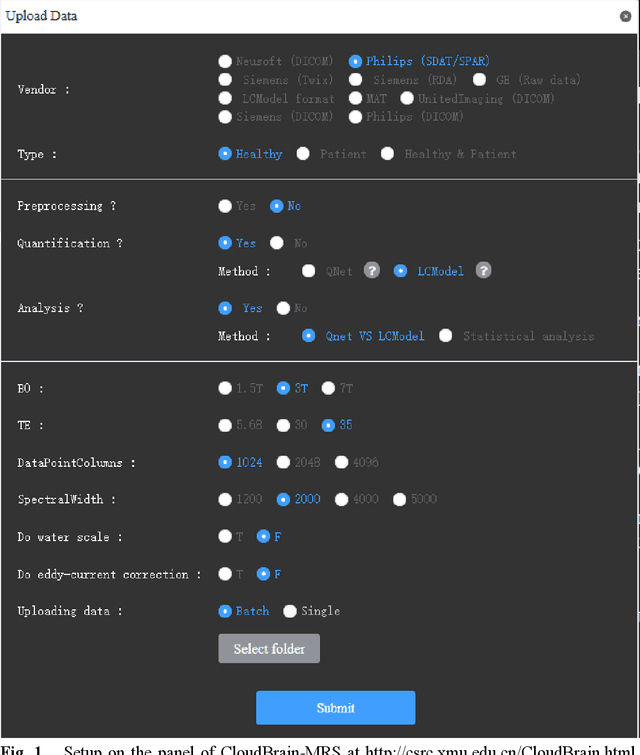
Abstract:Magnetic resonance spectroscopy (MRS) is an important clinical imaging method for the diagnosis of diseases. Spectrum is used to observe the signal intensity of metabolites or further infer their concentrations. Although the magnetic resonance vendors commonly provide basic functions of spectra plots and metabolite quantification, the widespread clinical research of MRS is still limited due to the lack of easy-to-use processing software or platform. To address this issue, we have developed CloudBrain-MRS, a cloud-based online platform that provides powerful hardware and advanced algorithms. The platform can be accessed simply through a web browser, without requiring any program installation on the user side. CloudBrain-MRS also integrates the classic LCModel and advanced artificial intelligence algorithms and supports batch preprocessing, quantification, and analysis of MRS data. Additionally, the platform offers useful functions: 1) Automatically statistical analysis to find biomarkers from the health and patient groups; 2) Consistency verification between the classic and artificial intelligence quantification algorithms; 3) Colorful and three-dimensional visualization for the easy observation of individual metabolite spectrum. Last, both healthy and mild cognitive impairment patient data are used to demonstrate the usefulness of the platform. To the best of our knowledge, this is the first cloud computing platform for in vivo MRS with artificial intelligence processing. We sincerely hope that this platform will facilitate efficient clinical research for MRS. CloudBrain-MRS is open-accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
Magnetic Resonance Spectroscopy Quantification Aided by Deep Estimations of Imperfection Factors and Overall Macromolecular Signal
Jun 16, 2023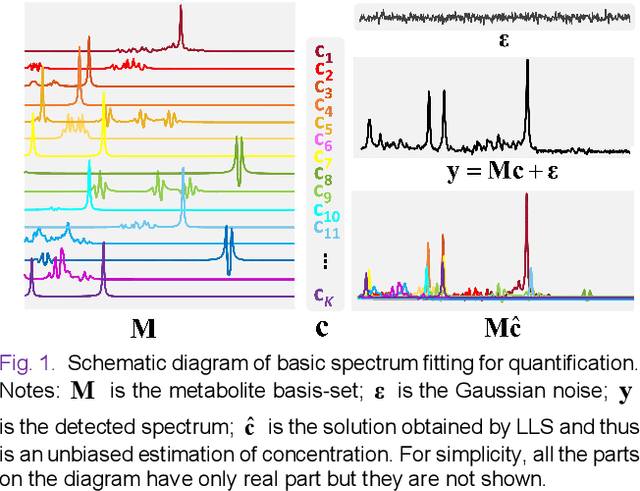
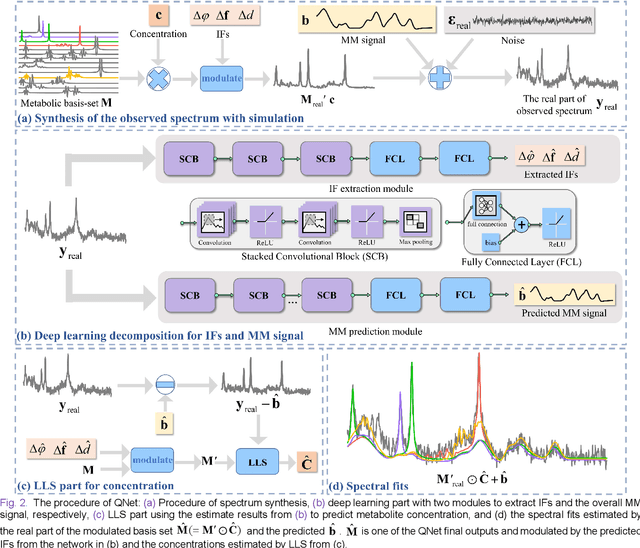
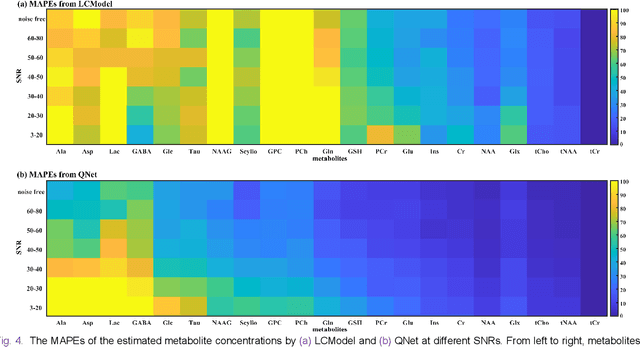
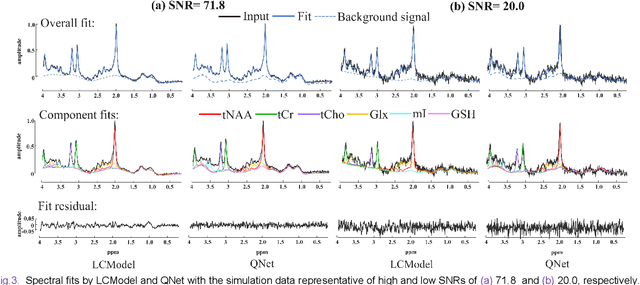
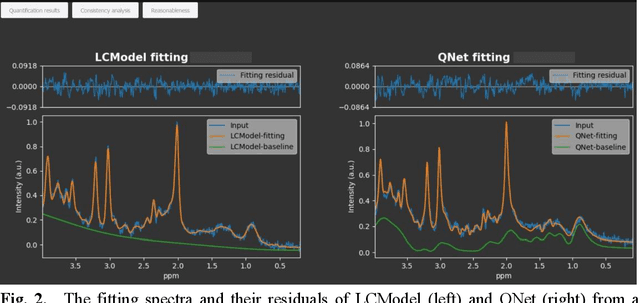
Abstract:Magnetic Resonance Spectroscopy (MRS) is an important non-invasive technique for in vivo biomedical detection. However, it is still challenging to accurately quantify metabolites with proton MRS due to three problems: Serious overlaps of metabolite signals, signal distortions due to non-ideal acquisition conditions and interference with strong background signals including macromolecule signals. The most popular software, LCModel, adopts the non-linear least square to quantify metabolites and addresses these problems by introducing regularization terms, imperfection factors of non-ideal acquisition conditions, and designing several empirical priors such as basissets of both metabolites and macromolecules. However, solving such a large non-linear quantitative problem is complicated. Moreover, when the signal-to-noise ratio of an input MRS signal is low, the solution may have a large deviation. In this work, deep learning is introduced to reduce the complexity of solving this overall quantitative problem. Deep learning is designed to predict directly the imperfection factors and the overall signal from macromolecules. Then, the remaining part of the quantification problem becomes a much simpler effective fitting and is easily solved by Linear Least Squares (LLS), which greatly improves the generalization to unseen concentration of metabolites in the training data. Experimental results show that compared with LCModel, the proposed method has smaller quantification errors for 700 sets of simulated test data, and presents more stable quantification results for 20 sets of healthy in vivo data at a wide range of signal-to-noise ratio. Qnet also outperforms other deep learning methods in terms of lower quantification error on most metabolites. Finally, QNet has been deployed on a cloud computing platform, CloudBrain-MRS, which is open accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
A Dynamics Theory of Implicit Regularization in Deep Low-Rank Matrix Factorization
Dec 29, 2022Abstract:Implicit regularization is an important way to interpret neural networks. Recent theory starts to explain implicit regularization with the model of deep matrix factorization (DMF) and analyze the trajectory of discrete gradient dynamics in the optimization process. These discrete gradient dynamics are relatively small but not infinitesimal, thus fitting well with the practical implementation of neural networks. Currently, discrete gradient dynamics analysis has been successfully applied to shallow networks but encounters the difficulty of complex computation for deep networks. In this work, we introduce another discrete gradient dynamics approach to explain implicit regularization, i.e. landscape analysis. It mainly focuses on gradient regions, such as saddle points and local minima. We theoretically establish the connection between saddle point escaping (SPE) stages and the matrix rank in DMF. We prove that, for a rank-R matrix reconstruction, DMF will converge to a second-order critical point after R stages of SPE. This conclusion is further experimentally verified on a low-rank matrix reconstruction problem. This work provides a new theory to analyze implicit regularization in deep learning.
Denoising Single Voxel Magnetic Resonance Spectroscopy with Deep Learning on Repeatedly Sampled In Vivo Data
Jan 26, 2021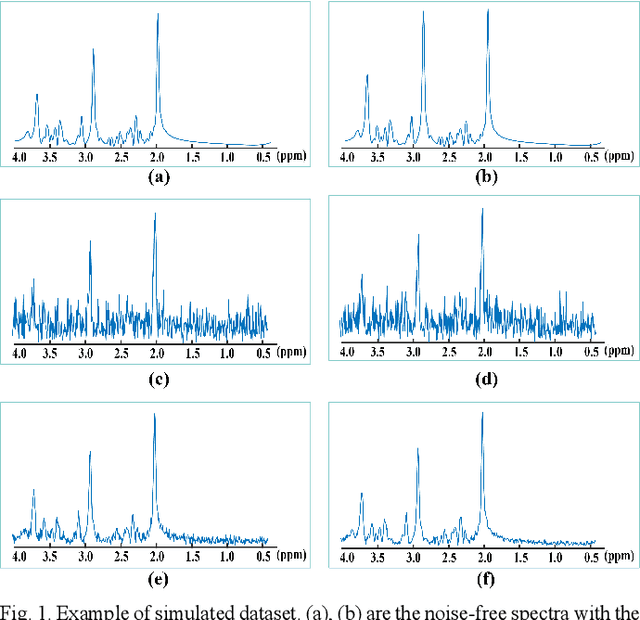

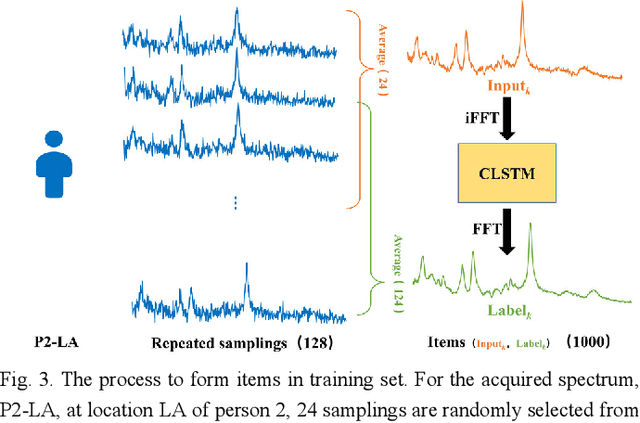
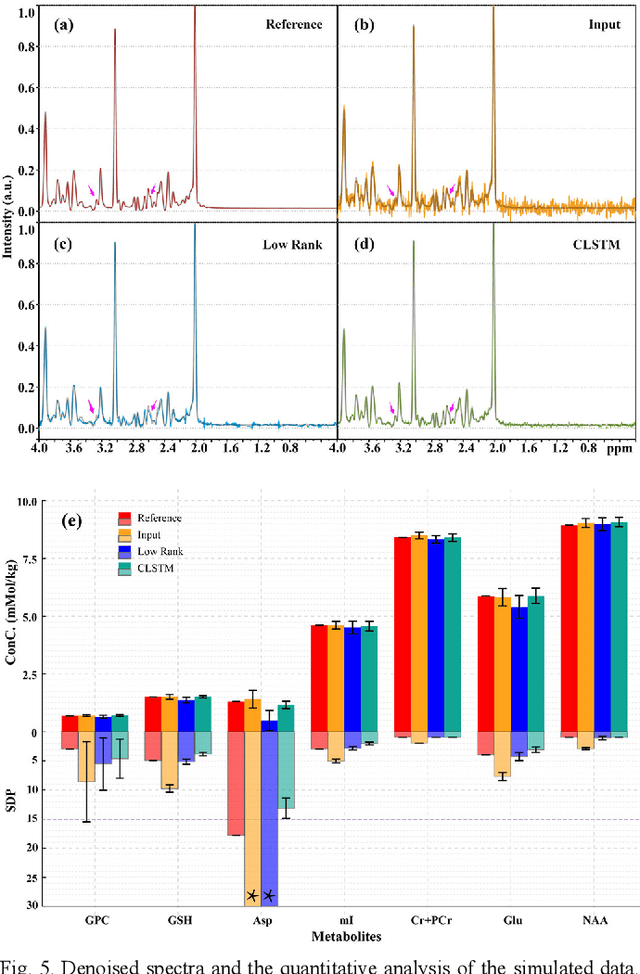
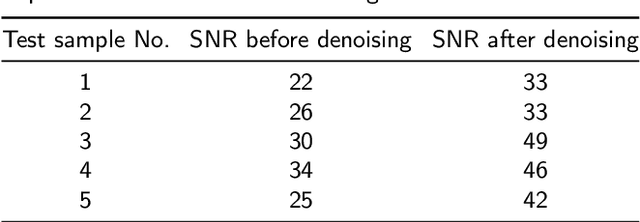
Abstract:Objective: Magnetic Resonance Spectroscopy (MRS) is a noninvasive tool to reveal metabolic information. One challenge of MRS is the relatively low Signal-Noise Ratio (SNR) due to low concentrations of metabolites. To improve the SNR, the most common approach is to average signals that are acquired in multiple times. The data acquisition time, however, is increased by multiple times accordingly, resulting in the scanned objects uncomfortable or even unbearable. Methods: By exploring the multiple sampled data, a deep learning denoising approach is proposed to learn a mapping from the low SNR signal to the high SNR one. Results: Results on simulated and in vivo data show that the proposed method significantly reduces the data acquisition time with slightly compromised metabolic accuracy. Conclusion: A deep learning denoising method was proposed to significantly shorten the time of data acquisition, while maintaining signal accuracy and reliability. Significance: Provide a solution of the fundamental low SNR problem in MRS with artificial intelligence.
Review and Prospect: Deep Learning in Nuclear Magnetic Resonance Spectroscopy
Jan 13, 2020



Abstract:Since the concept of deep learning (DL) was formally proposed in 2006, it had a major impact on academic research and industry. Nowadays, DL provides an unprecedented way to analyze and process data with demonstrated great results in computer vision, medical imaging, natural language processing, etc. In this Minireview, we summarize applications of DL in nuclear magnetic resonance (NMR) spectroscopy and outline a perspective for DL as entirely new approaches that is likely to transform NMR spectroscopy into a much more efficient and powerful technique in chemistry and life science.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge