Tianyu Qiu
Efficiently Solving Mixed-Hierarchy Games with Quasi-Policy Approximations
Feb 02, 2026Abstract:Multi-robot coordination often exhibits hierarchical structure, with some robots' decisions depending on the planned behaviors of others. While game theory provides a principled framework for such interactions, existing solvers struggle to handle mixed information structures that combine simultaneous (Nash) and hierarchical (Stackelberg) decision-making. We study N-robot forest-structured mixed-hierarchy games, in which each robot acts as a Stackelberg leader over its subtree while robots in different branches interact via Nash equilibria. We derive the Karush-Kuhn-Tucker (KKT) first-order optimality conditions for this class of games and show that they involve increasingly high-order derivatives of robots' best-response policies as the hierarchy depth grows, rendering a direct solution intractable. To overcome this challenge, we introduce a quasi-policy approximation that removes higher-order policy derivatives and develop an inexact Newton method for efficiently solving the resulting approximated KKT systems. We prove local exponential convergence of the proposed algorithm for games with non-quadratic objectives and nonlinear constraints. The approach is implemented in a highly optimized Julia library (MixedHierarchyGames.jl) and evaluated in simulated experiments, demonstrating real-time convergence for complex mixed-hierarchy information structures.
PSN Game: Game-theoretic Planning via a Player Selection Network
Apr 30, 2025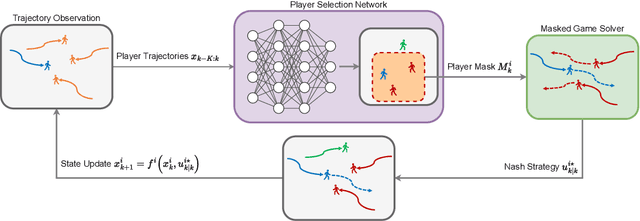
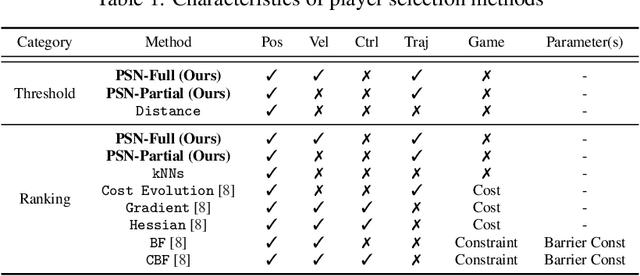
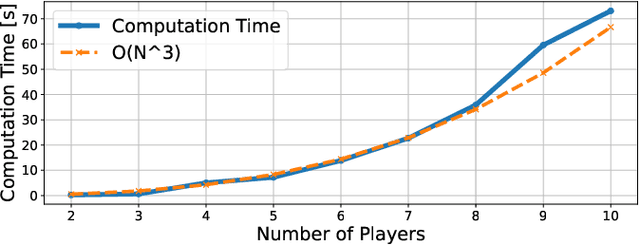
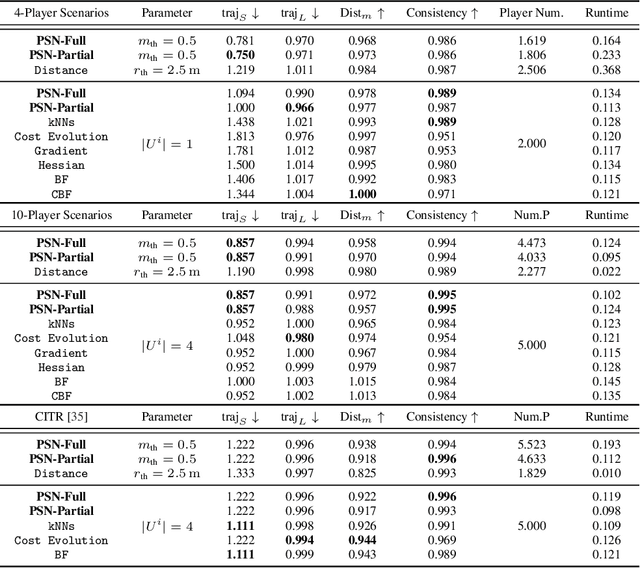
Abstract:While game-theoretic planning frameworks are effective at modeling multi-agent interactions, they require solving optimization problems with hundreds or thousands of variables, resulting in long computation times that limit their use in large-scale, real-time systems. To address this issue, we propose PSN Game: a novel game-theoretic planning framework that reduces runtime by learning a Player Selection Network (PSN). A PSN outputs a player selection mask that distinguishes influential players from less relevant ones, enabling the ego player to solve a smaller, masked game involving only selected players. By reducing the number of variables in the optimization problem, PSN directly lowers computation time. The PSN Game framework is more flexible than existing player selection methods as it i) relies solely on observations of players' past trajectories, without requiring full state, control, or other game-specific information; and ii) requires no online parameter tuning. We train PSNs in an unsupervised manner using a differentiable dynamic game solver, with reference trajectories from full-player games guiding the learning. Experiments in both simulated scenarios and human trajectory datasets demonstrate that i) PSNs outperform baseline selection methods in trajectory smoothness and length, while maintaining comparable safety and achieving a 10x speedup in runtime; and ii) PSNs generalize effectively to real-world scenarios without fine-tuning. By selecting only the most relevant players for decision-making, PSNs offer a general mechanism for reducing planning complexity that can be seamlessly integrated into existing multi-agent planning frameworks.
Enhancing Masked Time-Series Modeling via Dropping Patches
Dec 19, 2024



Abstract:This paper explores how to enhance existing masked time-series modeling by randomly dropping sub-sequence level patches of time series. On this basis, a simple yet effective method named DropPatch is proposed, which has two remarkable advantages: 1) It improves the pre-training efficiency by a square-level advantage; 2) It provides additional advantages for modeling in scenarios such as in-domain, cross-domain, few-shot learning and cold start. This paper conducts comprehensive experiments to verify the effectiveness of the method and analyze its internal mechanism. Empirically, DropPatch strengthens the attention mechanism, reduces information redundancy and serves as an efficient means of data augmentation. Theoretically, it is proved that DropPatch slows down the rate at which the Transformer representations collapse into the rank-1 linear subspace by randomly dropping patches, thus optimizing the quality of the learned representations
Dense Dynamics-Aware Reward Synthesis: Integrating Prior Experience with Demonstrations
Dec 02, 2024



Abstract:Many continuous control problems can be formulated as sparse-reward reinforcement learning (RL) tasks. In principle, online RL methods can automatically explore the state space to solve each new task. However, discovering sequences of actions that lead to a non-zero reward becomes exponentially more difficult as the task horizon increases. Manually shaping rewards can accelerate learning for a fixed task, but it is an arduous process that must be repeated for each new environment. We introduce a systematic reward-shaping framework that distills the information contained in 1) a task-agnostic prior data set and 2) a small number of task-specific expert demonstrations, and then uses these priors to synthesize dense dynamics-aware rewards for the given task. This supervision substantially accelerates learning in our experiments, and we provide analysis demonstrating how the approach can effectively guide online learning agents to faraway goals.
An Efficient and Generalizable Symbolic Regression Method for Time Series Analysis
Sep 06, 2024


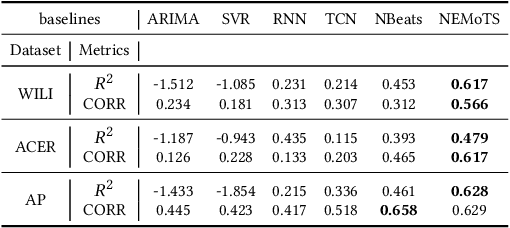
Abstract:Time series analysis and prediction methods currently excel in quantitative analysis, offering accurate future predictions and diverse statistical indicators, but generally falling short in elucidating the underlying evolution patterns of time series. To gain a more comprehensive understanding and provide insightful explanations, we utilize symbolic regression techniques to derive explicit expressions for the non-linear dynamics in the evolution of time series variables. However, these techniques face challenges in computational efficiency and generalizability across diverse real-world time series data. To overcome these challenges, we propose \textbf{N}eural-\textbf{E}nhanced \textbf{Mo}nte-Carlo \textbf{T}ree \textbf{S}earch (NEMoTS) for time series. NEMoTS leverages the exploration-exploitation balance of Monte-Carlo Tree Search (MCTS), significantly reducing the search space in symbolic regression and improving expression quality. Furthermore, by integrating neural networks with MCTS, NEMoTS not only capitalizes on their superior fitting capabilities to concentrate on more pertinent operations post-search space reduction, but also replaces the complex and time-consuming simulation process, thereby substantially improving computational efficiency and generalizability in time series analysis. NEMoTS offers an efficient and comprehensive approach to time series analysis. Experiments with three real-world datasets demonstrate NEMoTS's significant superiority in performance, efficiency, reliability, and interpretability, making it well-suited for large-scale real-world time series data.
CloudBrain-NMR: An Intelligent Cloud Computing Platform for NMR Spectroscopy Processing, Reconstruction and Analysis
Sep 12, 2023



Abstract:Nuclear Magnetic Resonance (NMR) spectroscopy has served as a powerful analytical tool for studying molecular structure and dynamics in chemistry and biology. However, the processing of raw data acquired from NMR spectrometers and subsequent quantitative analysis involves various specialized tools, which necessitates comprehensive knowledge in programming and NMR. Particularly, the emerging deep learning tools is hard to be widely used in NMR due to the sophisticated setup of computation. Thus, NMR processing is not an easy task for chemist and biologists. In this work, we present CloudBrain-NMR, an intelligent online cloud computing platform designed for NMR data reading, processing, reconstruction, and quantitative analysis. The platform is conveniently accessed through a web browser, eliminating the need for any program installation on the user side. CloudBrain-NMR uses parallel computing with graphics processing units and central processing units, resulting in significantly shortened computation time. Furthermore, it incorporates state-of-the-art deep learning-based algorithms offering comprehensive functionalities that allow users to complete the entire processing procedure without relying on additional software. This platform has empowered NMR applications with advanced artificial intelligence processing. CloudBrain-NMR is openly accessible for free usage at https://csrc.xmu.edu.cn/CloudBrain.html
Resolution enhancement of NMR by decoupling with low-rank Hankel model
Dec 02, 2022

Abstract:Nuclear magnetic resonance (NMR) spectroscopy has become a formidable tool for biochemistry and medicine. Although J-coupling carries essential structural information it may also limit the spectral resolution. Homonuclear decoupling remains a challenging problem. In this work, we introduce a new approach that uses a specific coupling value as prior knowledge, and Hankel property of exponential NMR signal to achieve the broadband heteronuclear decoupling using the low-rank method. Our results on synthetic and realistic HMQC spectra demonstrate that the proposed method not only effectively enhances resolution by decoupling, but also maintains sensitivity and suppresses spectral artefacts. The approach can be combined with the non-uniform sampling, which means that the resolution can be further improved without any extra acquisition time
On the Effectiveness of Sampled Softmax Loss for Item Recommendation
Jan 07, 2022

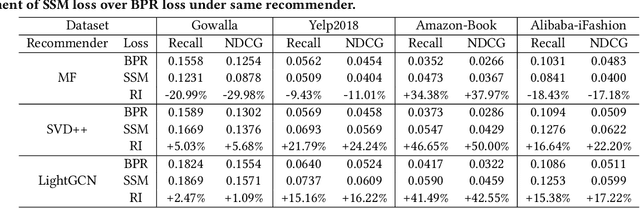
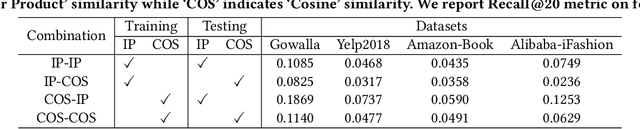
Abstract:Learning objectives of recommender models remain largely unexplored. Most methods routinely adopt either pointwise or pairwise loss to train the model parameters, while rarely pay attention to softmax loss due to the high computational cost. Sampled softmax loss emerges as an efficient substitute for softmax loss. Its special case, InfoNCE loss, has been widely used in self-supervised learning and exhibited remarkable performance for contrastive learning. Nonetheless, limited studies use sampled softmax loss as the learning objective to train the recommender. Worse still, none of them explore its properties and answer "Does sampled softmax loss suit for item recommendation?" and "What are the conceptual advantages of sampled softmax loss, as compared with the prevalent losses?", to the best of our knowledge. In this work, we aim to better understand sampled softmax loss for item recommendation. Specifically, we first theoretically reveal three model-agnostic advantages: (1) mitigating popularity bias, which is beneficial to long-tail recommendation; (2) mining hard negative samples, which offers informative gradients to optimize model parameters; and (3) maximizing the ranking metric, which facilitates top-K performance. Moreover, we probe the model-specific characteristics on the top of various recommenders. Experimental results suggest that sampled softmax loss is more friendly to history and graph-based recommenders (e.g., SVD++ and LightGCN), but performs poorly for ID-based models (e.g., MF). We ascribe this to its shortcoming in learning representation magnitude, making the combination with the models that are also incapable of adjusting representation magnitude learn poor representations. In contrast, the history- and graph-based models, which naturally adjust representation magnitude according to node degree, are able to compensate for the shortcoming of sampled softmax loss.
Denoising Single Voxel Magnetic Resonance Spectroscopy with Deep Learning on Repeatedly Sampled In Vivo Data
Jan 26, 2021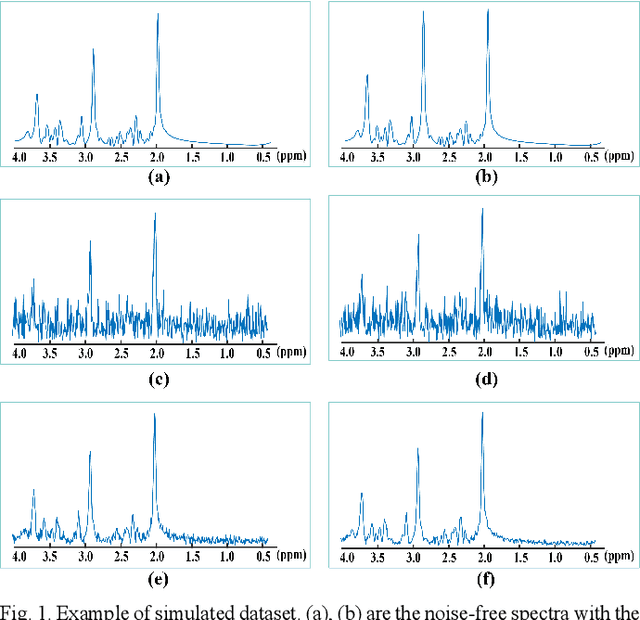

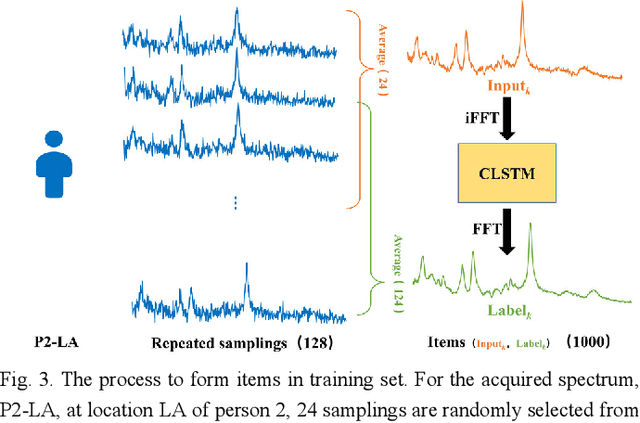
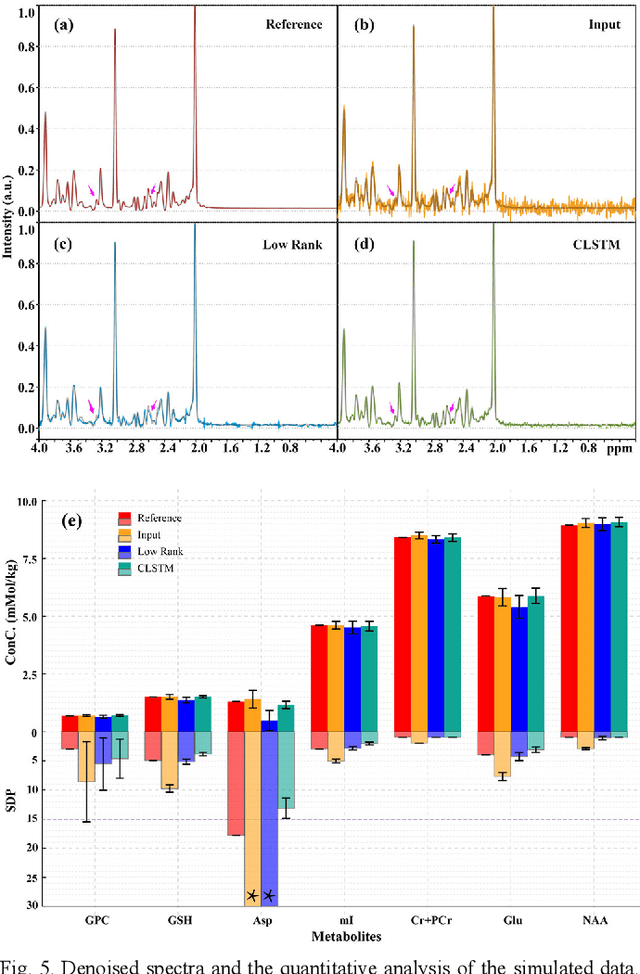
Abstract:Objective: Magnetic Resonance Spectroscopy (MRS) is a noninvasive tool to reveal metabolic information. One challenge of MRS is the relatively low Signal-Noise Ratio (SNR) due to low concentrations of metabolites. To improve the SNR, the most common approach is to average signals that are acquired in multiple times. The data acquisition time, however, is increased by multiple times accordingly, resulting in the scanned objects uncomfortable or even unbearable. Methods: By exploring the multiple sampled data, a deep learning denoising approach is proposed to learn a mapping from the low SNR signal to the high SNR one. Results: Results on simulated and in vivo data show that the proposed method significantly reduces the data acquisition time with slightly compromised metabolic accuracy. Conclusion: A deep learning denoising method was proposed to significantly shorten the time of data acquisition, while maintaining signal accuracy and reliability. Significance: Provide a solution of the fundamental low SNR problem in MRS with artificial intelligence.
Accelerated Nuclear Magnetic Resonance Spectroscopy with Deep Learning
May 14, 2019
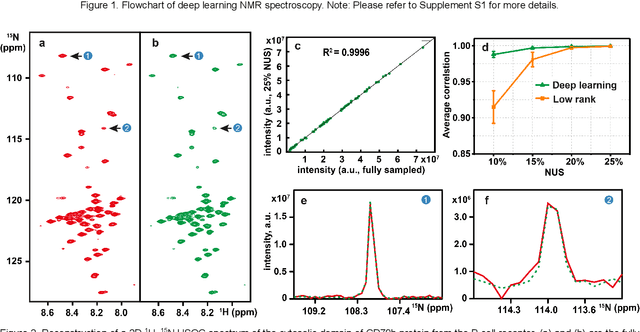
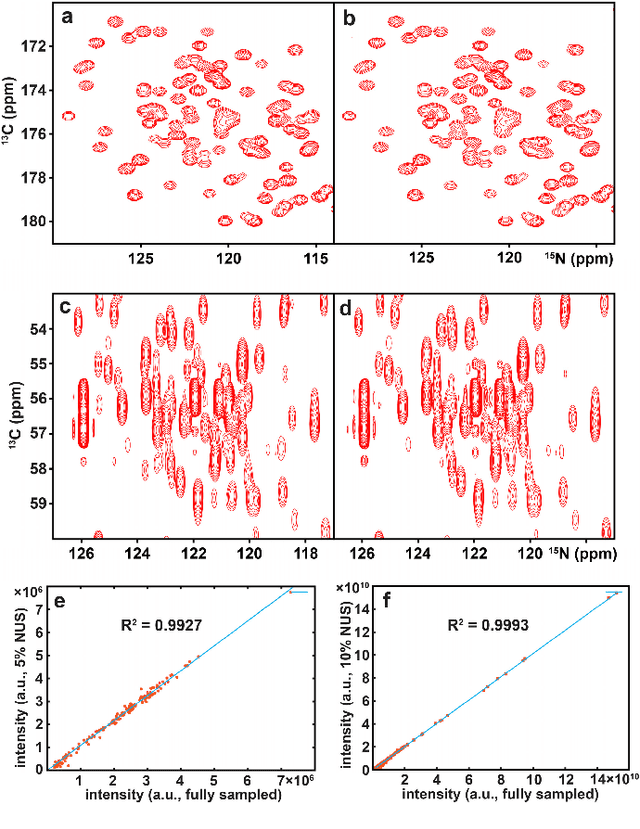

Abstract:Nuclear magnetic resonance (NMR) spectroscopy serves as an indispensable tool in chemistry and biology but often suffers from long experimental time. We present a proof-of-concept of application of deep learning and neural network for high-quality, reliable, and very fast NMR spectra reconstruction from limited experimental data. We show that the neural network training can be achieved using solely synthetic NMR signal, which lifts the prohibiting demand for a large volume of realistic training data usually required in the deep learning approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge