Zhangren Tu
An artificially intelligent magnetic resonance spectroscopy quantification method: Comparison between QNet and LCModel on the cloud computing platform CloudBrain-MRS
Mar 06, 2025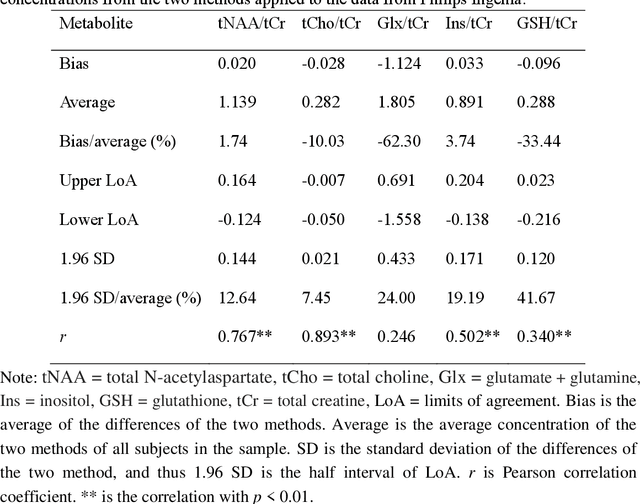
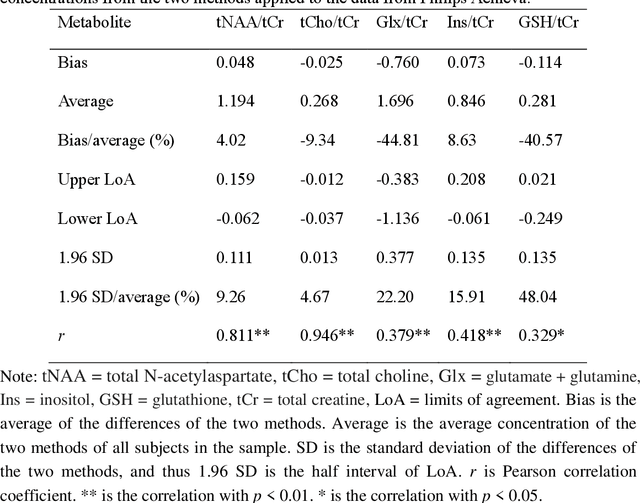
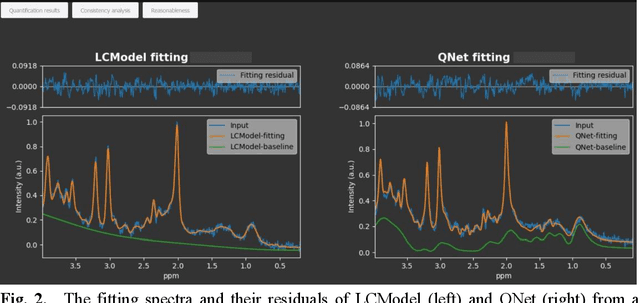
Abstract:Objctives: This work aimed to statistically compare the metabolite quantification of human brain magnetic resonance spectroscopy (MRS) between the deep learning method QNet and the classical method LCModel through an easy-to-use intelligent cloud computing platform CloudBrain-MRS. Materials and Methods: In this retrospective study, two 3 T MRI scanners Philips Ingenia and Achieva collected 61 and 46 in vivo 1H magnetic resonance (MR) spectra of healthy participants, respectively, from the brain region of pregenual anterior cingulate cortex from September to October 2021. The analyses of Bland-Altman, Pearson correlation and reasonability were performed to assess the degree of agreement, linear correlation and reasonability between the two quantification methods. Results: Fifteen healthy volunteers (12 females and 3 males, age range: 21-35 years, mean age/standard deviation = 27.4/3.9 years) were recruited. The analyses of Bland-Altman, Pearson correlation and reasonability showed high to good consistency and very strong to moderate correlation between the two methods for quantification of total N-acetylaspartate (tNAA), total choline (tCho), and inositol (Ins) (relative half interval of limits of agreement = 3.04%, 9.3%, and 18.5%, respectively; Pearson correlation coefficient r = 0.775, 0.927, and 0.469, respectively). In addition, quantification results of QNet are more likely to be closer to the previous reported average values than those of LCModel. Conclusion: There were high or good degrees of consistency between the quantification results of QNet and LCModel for tNAA, tCho, and Ins, and QNet generally has more reasonable quantification than LCModel.
CloudBrain-NMR: An Intelligent Cloud Computing Platform for NMR Spectroscopy Processing, Reconstruction and Analysis
Sep 12, 2023



Abstract:Nuclear Magnetic Resonance (NMR) spectroscopy has served as a powerful analytical tool for studying molecular structure and dynamics in chemistry and biology. However, the processing of raw data acquired from NMR spectrometers and subsequent quantitative analysis involves various specialized tools, which necessitates comprehensive knowledge in programming and NMR. Particularly, the emerging deep learning tools is hard to be widely used in NMR due to the sophisticated setup of computation. Thus, NMR processing is not an easy task for chemist and biologists. In this work, we present CloudBrain-NMR, an intelligent online cloud computing platform designed for NMR data reading, processing, reconstruction, and quantitative analysis. The platform is conveniently accessed through a web browser, eliminating the need for any program installation on the user side. CloudBrain-NMR uses parallel computing with graphics processing units and central processing units, resulting in significantly shortened computation time. Furthermore, it incorporates state-of-the-art deep learning-based algorithms offering comprehensive functionalities that allow users to complete the entire processing procedure without relying on additional software. This platform has empowered NMR applications with advanced artificial intelligence processing. CloudBrain-NMR is openly accessible for free usage at https://csrc.xmu.edu.cn/CloudBrain.html
CloudBrain-MRS: An Intelligent Cloud Computing Platform for in vivo Magnetic Resonance Spectroscopy Preprocessing, Quantification, and Analysis
Jun 19, 2023
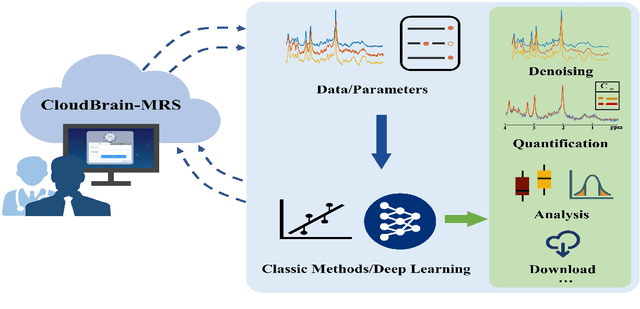
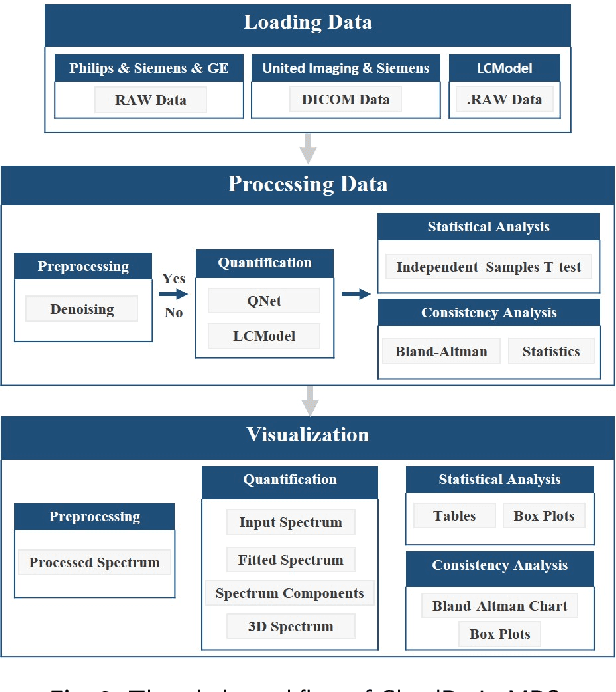
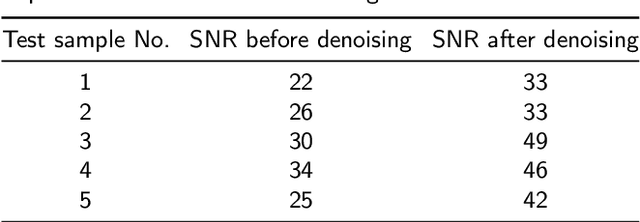
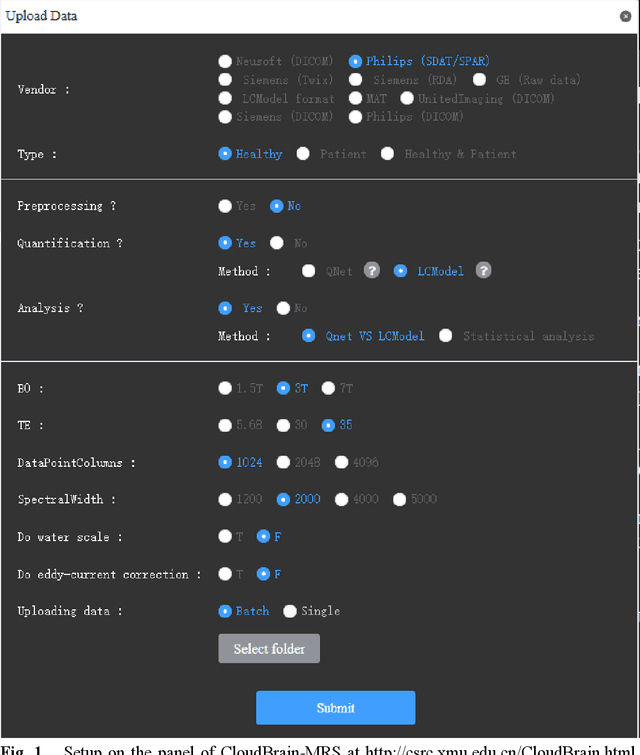
Abstract:Magnetic resonance spectroscopy (MRS) is an important clinical imaging method for the diagnosis of diseases. Spectrum is used to observe the signal intensity of metabolites or further infer their concentrations. Although the magnetic resonance vendors commonly provide basic functions of spectra plots and metabolite quantification, the widespread clinical research of MRS is still limited due to the lack of easy-to-use processing software or platform. To address this issue, we have developed CloudBrain-MRS, a cloud-based online platform that provides powerful hardware and advanced algorithms. The platform can be accessed simply through a web browser, without requiring any program installation on the user side. CloudBrain-MRS also integrates the classic LCModel and advanced artificial intelligence algorithms and supports batch preprocessing, quantification, and analysis of MRS data. Additionally, the platform offers useful functions: 1) Automatically statistical analysis to find biomarkers from the health and patient groups; 2) Consistency verification between the classic and artificial intelligence quantification algorithms; 3) Colorful and three-dimensional visualization for the easy observation of individual metabolite spectrum. Last, both healthy and mild cognitive impairment patient data are used to demonstrate the usefulness of the platform. To the best of our knowledge, this is the first cloud computing platform for in vivo MRS with artificial intelligence processing. We sincerely hope that this platform will facilitate efficient clinical research for MRS. CloudBrain-MRS is open-accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
Alternating Deep Low Rank Approach for Exponential Function Reconstruction and Its Biomedical Magnetic Resonance Applications
Nov 24, 2022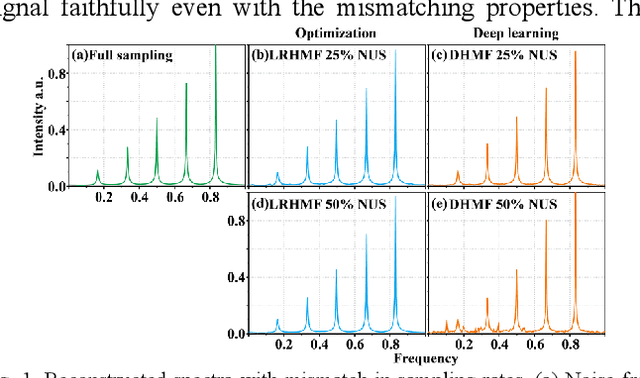
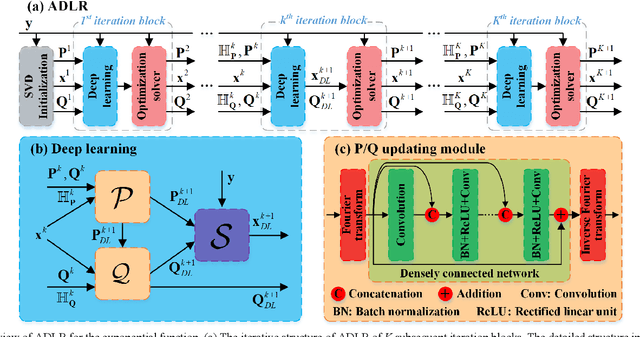
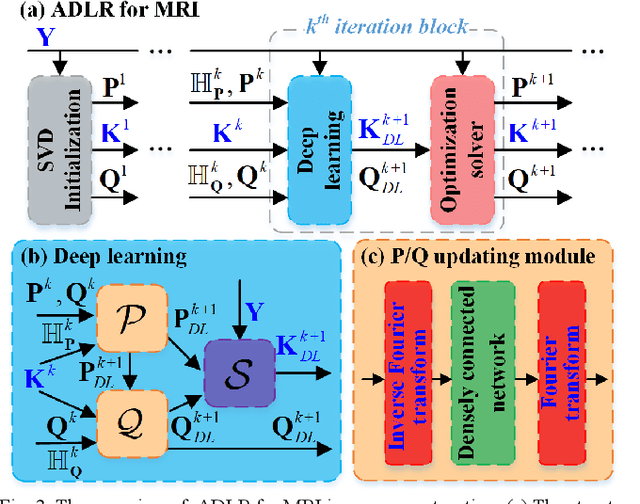
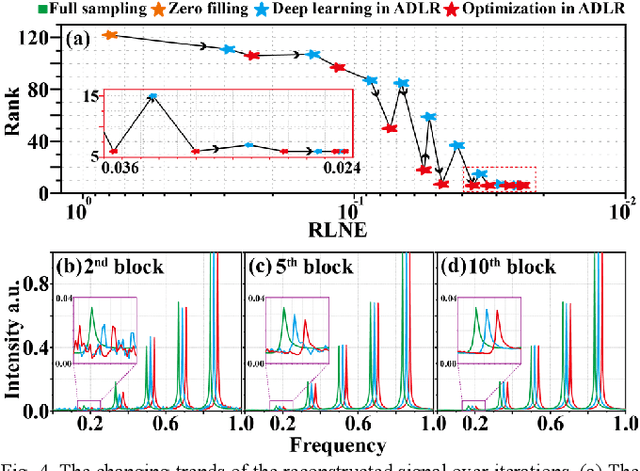
Abstract:Exponential function is a fundamental signal form in general signal processing and biomedical applications, such as magnetic resonance spectroscopy and imaging. How to reduce the sampling time of these signals is an important problem. Sub-Nyquist sampling can accelerate signal acquisition but bring in artifacts. Recently, the low rankness of these exponentials has been applied to implicitly constrain the deep learning network through the unrolling of low rank Hankel factorization algorithm. However, only depending on the implicit low rank constraint cannot provide the robust reconstruction, such as sampling rate mismatches. In this work, by introducing the explicit low rank prior to constrain the deep learning, we propose an Alternating Deep Low Rank approach (ADLR) that utilizes deep learning and optimization solvers alternately. The former solver accelerates the reconstruction while the latter one corrects the reconstruction error from the mismatch. The experiments on both general exponential functions and realistic biomedical magnetic resonance data show that, compared with the state-of-the-art methods, ADLR can achieve much lower reconstruction error and effectively alleviates the decrease of reconstruction quality with sampling rate mismatches.
Accelerated NMR Spectroscopy: Merge Optimization with Deep Learning
Jan 02, 2021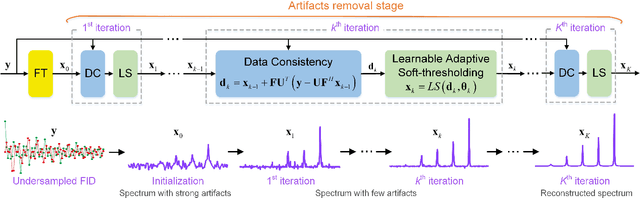
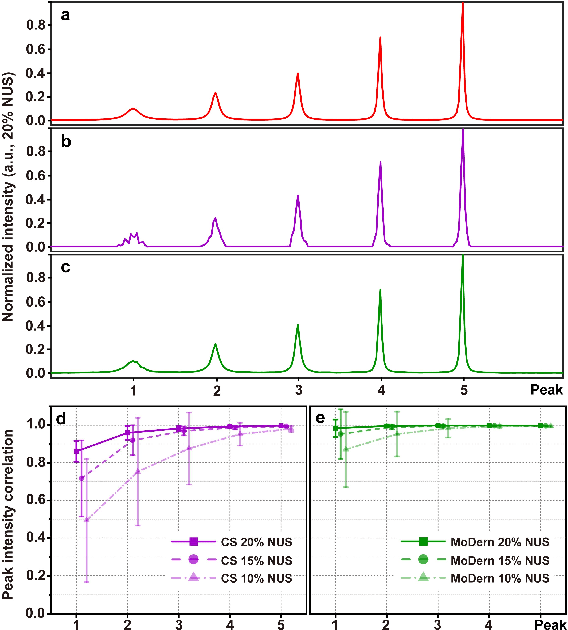
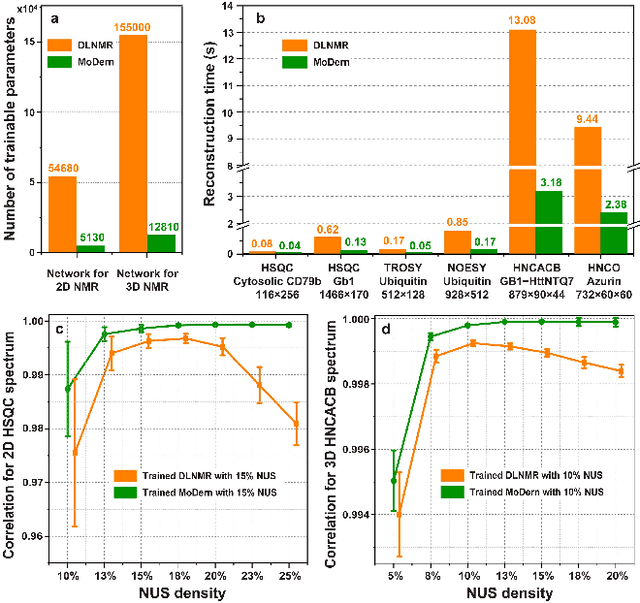
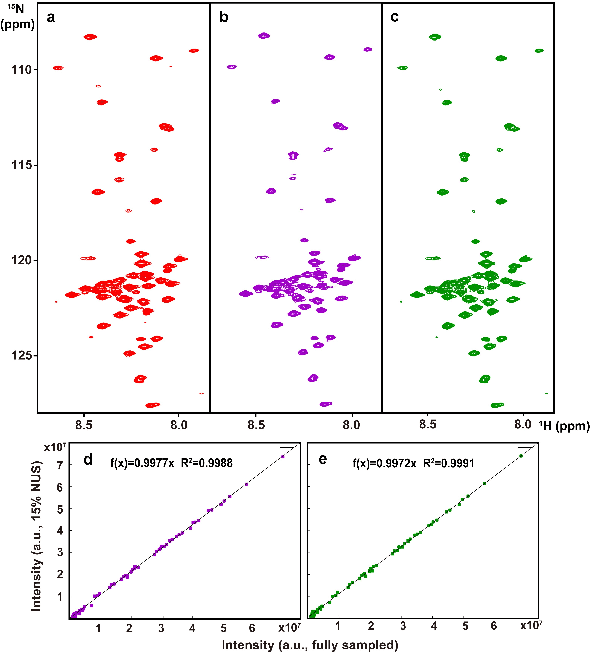
Abstract:Multi-dimensional NMR spectroscopy is an invaluable biophysical tool in studies of structure, interactions, and dynamics of large molecules like proteins and nuclear acids. Non-uniform sampling is a powerful approach for shortening measurement time and increasing spectra resolution. Several methods have been established for spectra reconstruction from the undersampled data, typical approaches include model-based optimization and data-driven deep learning. The former is well theoretically grounded and provides high-quality spectra, while the latter has a huge advantage in reconstruction time potential and push further limits of spectra quality and analysis. Combining the merits of the two, we propose a model-inspired deep learning, for reliable, high-quality, and ultra-fast spectra reconstruction, exemplified by multi-dimensional spectra of several representative proteins. We demonstrate that the proposed network needs very few parameters, and shows very high robustness in respect to dissimilarity of the training and target data in the spectra size, type, and sampling level. This work can be considered as a proof-of-concept of merging optimization with deep learning in NMR spectroscopy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge