Meijin Lin
An artificially intelligent magnetic resonance spectroscopy quantification method: Comparison between QNet and LCModel on the cloud computing platform CloudBrain-MRS
Mar 06, 2025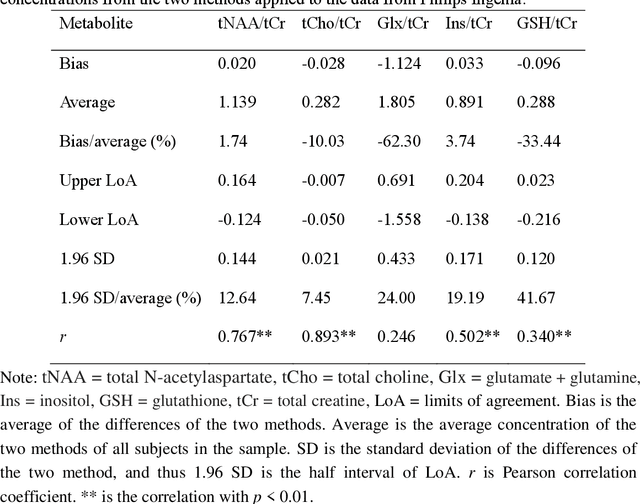
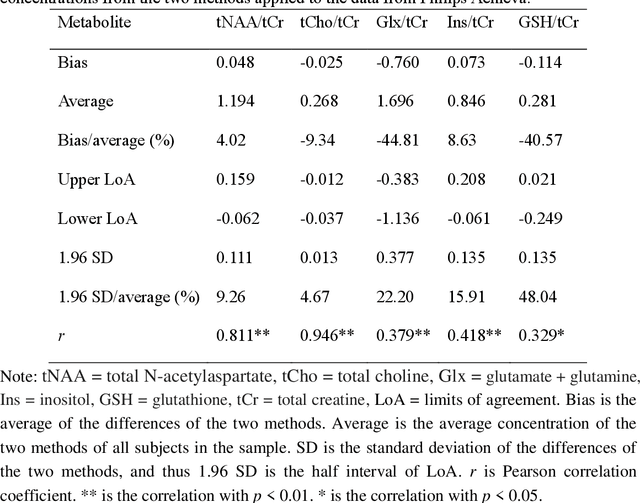
Abstract:Objctives: This work aimed to statistically compare the metabolite quantification of human brain magnetic resonance spectroscopy (MRS) between the deep learning method QNet and the classical method LCModel through an easy-to-use intelligent cloud computing platform CloudBrain-MRS. Materials and Methods: In this retrospective study, two 3 T MRI scanners Philips Ingenia and Achieva collected 61 and 46 in vivo 1H magnetic resonance (MR) spectra of healthy participants, respectively, from the brain region of pregenual anterior cingulate cortex from September to October 2021. The analyses of Bland-Altman, Pearson correlation and reasonability were performed to assess the degree of agreement, linear correlation and reasonability between the two quantification methods. Results: Fifteen healthy volunteers (12 females and 3 males, age range: 21-35 years, mean age/standard deviation = 27.4/3.9 years) were recruited. The analyses of Bland-Altman, Pearson correlation and reasonability showed high to good consistency and very strong to moderate correlation between the two methods for quantification of total N-acetylaspartate (tNAA), total choline (tCho), and inositol (Ins) (relative half interval of limits of agreement = 3.04%, 9.3%, and 18.5%, respectively; Pearson correlation coefficient r = 0.775, 0.927, and 0.469, respectively). In addition, quantification results of QNet are more likely to be closer to the previous reported average values than those of LCModel. Conclusion: There were high or good degrees of consistency between the quantification results of QNet and LCModel for tNAA, tCho, and Ins, and QNet generally has more reasonable quantification than LCModel.
Reproducibility Assessment of Magnetic Resonance Spectroscopy of Pregenual Anterior Cingulate Cortex across Sessions and Vendors via the Cloud Computing Platform CloudBrain-MRS
Mar 06, 2025



Abstract:Given the need to elucidate the mechanisms underlying illnesses and their treatment, as well as the lack of harmonization of acquisition and post-processing protocols among different magnetic resonance system vendors, this work is to determine if metabolite concentrations obtained from different sessions, machine models and even different vendors of 3 T scanners can be highly reproducible and be pooled for diagnostic analysis, which is very valuable for the research of rare diseases. Participants underwent magnetic resonance imaging (MRI) scanning once on two separate days within one week (one session per day, each session including two proton magnetic resonance spectroscopy (1H-MRS) scans with no more than a 5-minute interval between scans (no off-bed activity)) on each machine. were analyzed for reliability of within- and between- sessions using the coefficient of variation (CV) and intraclass correlation coefficient (ICC), and for reproducibility of across the machines using correlation coefficient. As for within- and between- session, all CV values for a group of all the first or second scans of a session, or for a session were almost below 20%, and most of the ICCs for metabolites range from moderate (0.4-0.59) to excellent (0.75-1), indicating high data reliability. When it comes to the reproducibility across the three scanners, all Pearson correlation coefficients across the three machines approached 1 with most around 0.9, and majority demonstrated statistical significance (P<0.01). Additionally, the intra-vendor reproducibility was greater than the inter-vendor ones.
NMR Spectra Denoising with Vandermonde Constraints
Oct 21, 2023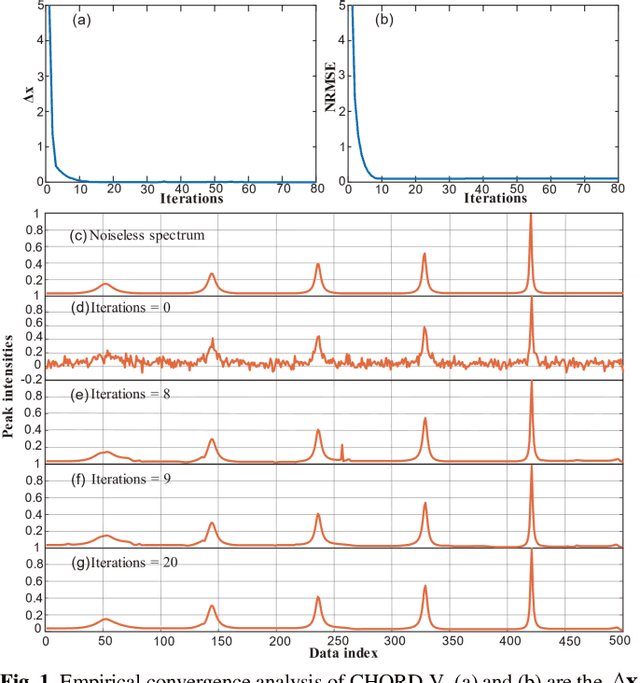
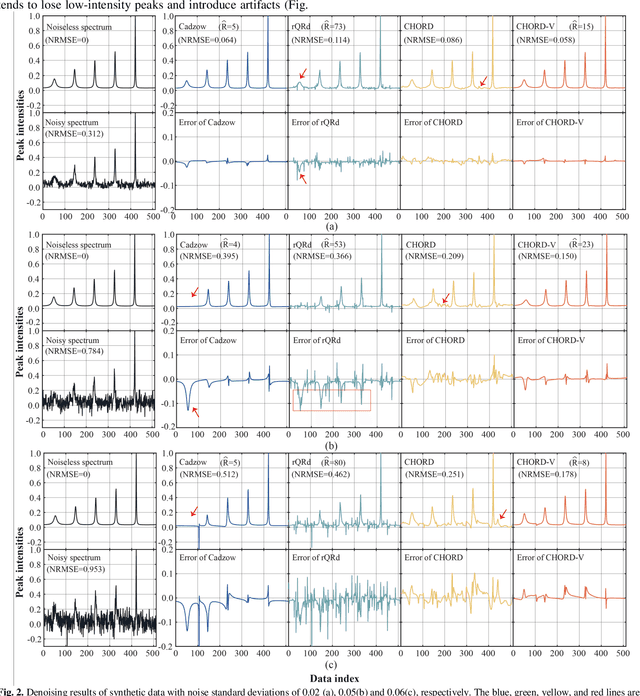
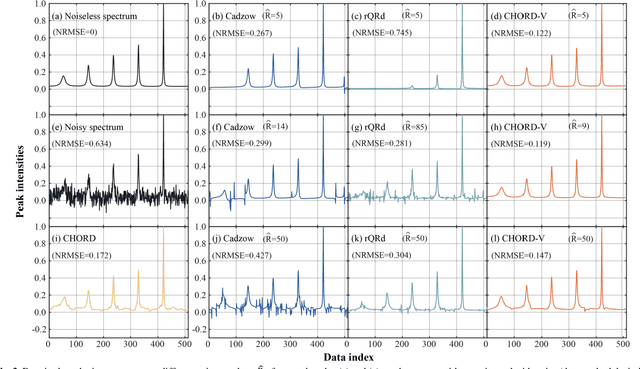

Abstract:Nuclear magnetic resonance (NMR) spectroscopy serves as an important tool to analyze chemicals and proteins in bioengineering. However, NMR signals are easily contaminated by noise during the data acquisition, which can affect subsequent quantitative analysis. Therefore, denoising NMR signals has been a long-time concern. In this work, we propose an optimization model-based iterative denoising method, CHORD-V, by treating the time-domain NMR signal as damped exponentials and maintaining the exponential signal form with a Vandermonde factorization. Results on both synthetic and realistic NMR data show that CHORD-V has a superior denoising performance over typical Cadzow and rQRd methods, and the state-of-the-art CHORD method. CHORD-V restores low-intensity spectral peaks more accurately, especially when the noise is relatively high.
CloudBrain-NMR: An Intelligent Cloud Computing Platform for NMR Spectroscopy Processing, Reconstruction and Analysis
Sep 12, 2023



Abstract:Nuclear Magnetic Resonance (NMR) spectroscopy has served as a powerful analytical tool for studying molecular structure and dynamics in chemistry and biology. However, the processing of raw data acquired from NMR spectrometers and subsequent quantitative analysis involves various specialized tools, which necessitates comprehensive knowledge in programming and NMR. Particularly, the emerging deep learning tools is hard to be widely used in NMR due to the sophisticated setup of computation. Thus, NMR processing is not an easy task for chemist and biologists. In this work, we present CloudBrain-NMR, an intelligent online cloud computing platform designed for NMR data reading, processing, reconstruction, and quantitative analysis. The platform is conveniently accessed through a web browser, eliminating the need for any program installation on the user side. CloudBrain-NMR uses parallel computing with graphics processing units and central processing units, resulting in significantly shortened computation time. Furthermore, it incorporates state-of-the-art deep learning-based algorithms offering comprehensive functionalities that allow users to complete the entire processing procedure without relying on additional software. This platform has empowered NMR applications with advanced artificial intelligence processing. CloudBrain-NMR is openly accessible for free usage at https://csrc.xmu.edu.cn/CloudBrain.html
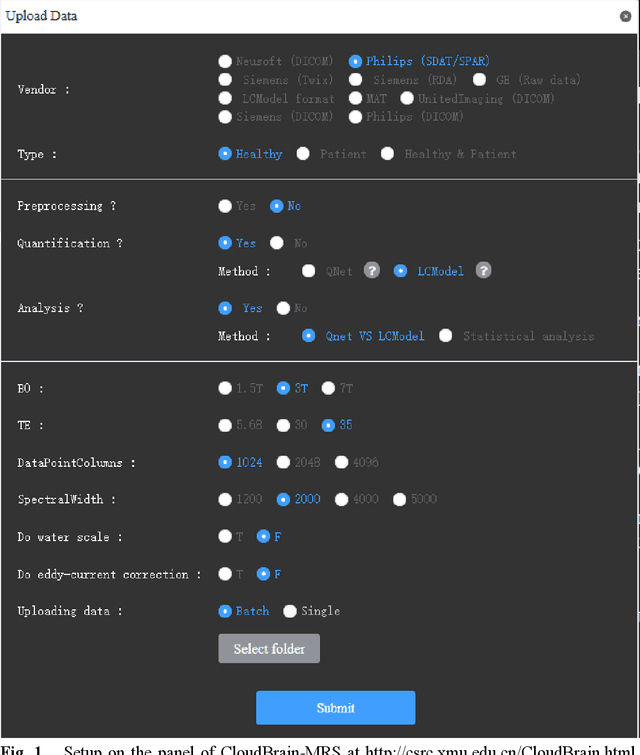
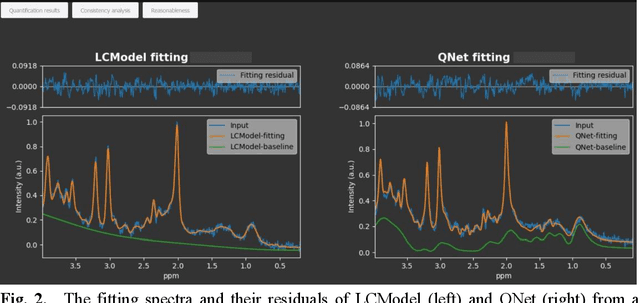
CloudBrain-MRS: An Intelligent Cloud Computing Platform for in vivo Magnetic Resonance Spectroscopy Preprocessing, Quantification, and Analysis
Jun 19, 2023
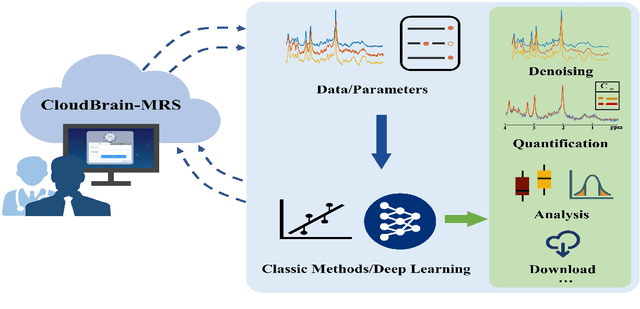
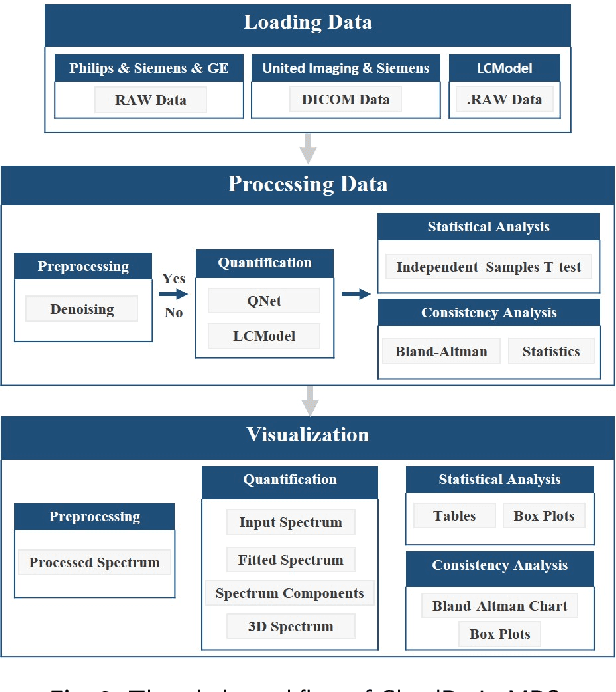
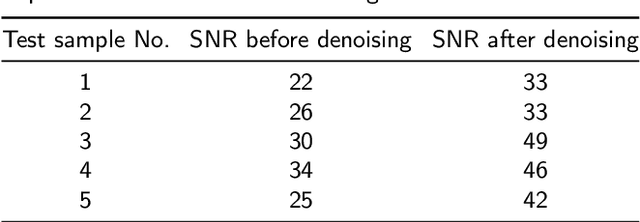
Abstract:Magnetic resonance spectroscopy (MRS) is an important clinical imaging method for the diagnosis of diseases. Spectrum is used to observe the signal intensity of metabolites or further infer their concentrations. Although the magnetic resonance vendors commonly provide basic functions of spectra plots and metabolite quantification, the widespread clinical research of MRS is still limited due to the lack of easy-to-use processing software or platform. To address this issue, we have developed CloudBrain-MRS, a cloud-based online platform that provides powerful hardware and advanced algorithms. The platform can be accessed simply through a web browser, without requiring any program installation on the user side. CloudBrain-MRS also integrates the classic LCModel and advanced artificial intelligence algorithms and supports batch preprocessing, quantification, and analysis of MRS data. Additionally, the platform offers useful functions: 1) Automatically statistical analysis to find biomarkers from the health and patient groups; 2) Consistency verification between the classic and artificial intelligence quantification algorithms; 3) Colorful and three-dimensional visualization for the easy observation of individual metabolite spectrum. Last, both healthy and mild cognitive impairment patient data are used to demonstrate the usefulness of the platform. To the best of our knowledge, this is the first cloud computing platform for in vivo MRS with artificial intelligence processing. We sincerely hope that this platform will facilitate efficient clinical research for MRS. CloudBrain-MRS is open-accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
Magnetic Resonance Spectroscopy Quantification Aided by Deep Estimations of Imperfection Factors and Overall Macromolecular Signal
Jun 16, 2023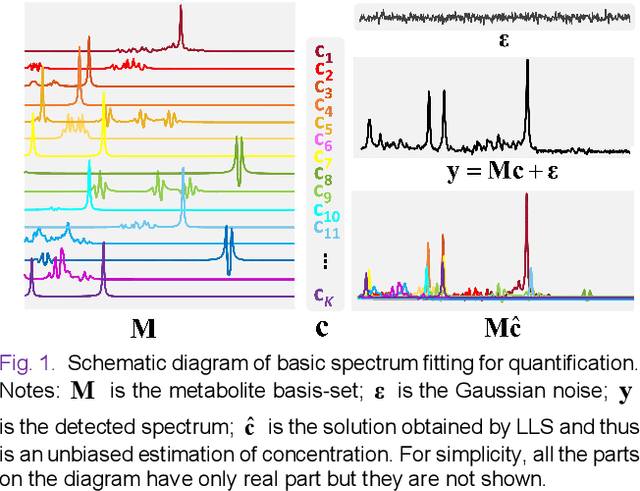
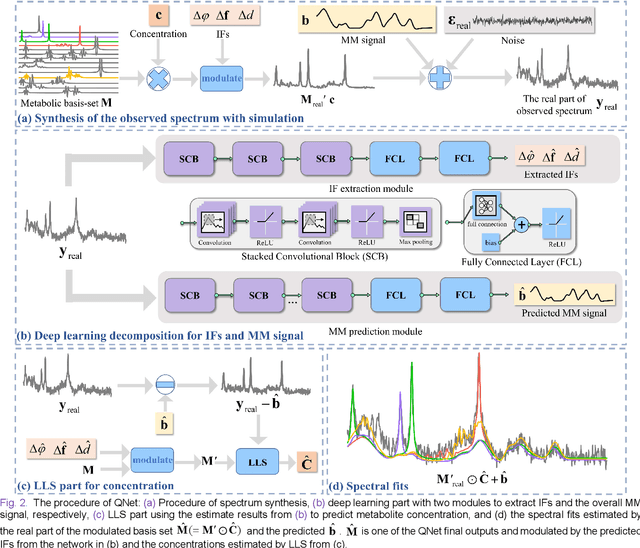
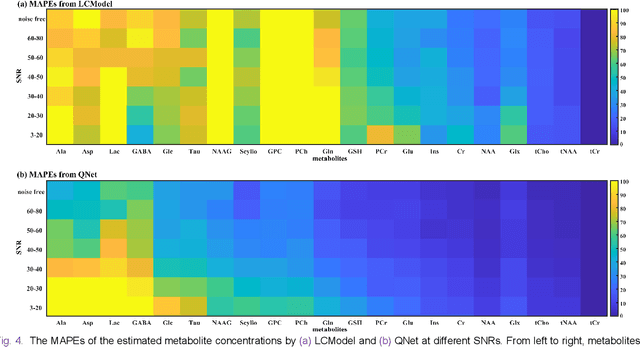
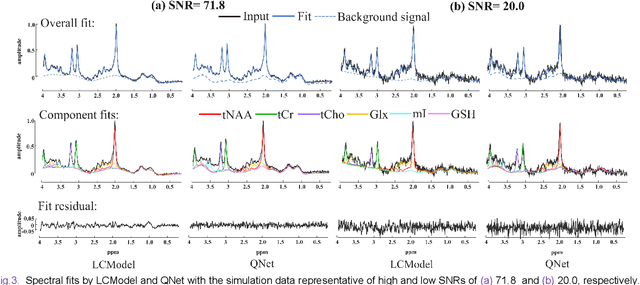
Abstract:Magnetic Resonance Spectroscopy (MRS) is an important non-invasive technique for in vivo biomedical detection. However, it is still challenging to accurately quantify metabolites with proton MRS due to three problems: Serious overlaps of metabolite signals, signal distortions due to non-ideal acquisition conditions and interference with strong background signals including macromolecule signals. The most popular software, LCModel, adopts the non-linear least square to quantify metabolites and addresses these problems by introducing regularization terms, imperfection factors of non-ideal acquisition conditions, and designing several empirical priors such as basissets of both metabolites and macromolecules. However, solving such a large non-linear quantitative problem is complicated. Moreover, when the signal-to-noise ratio of an input MRS signal is low, the solution may have a large deviation. In this work, deep learning is introduced to reduce the complexity of solving this overall quantitative problem. Deep learning is designed to predict directly the imperfection factors and the overall signal from macromolecules. Then, the remaining part of the quantification problem becomes a much simpler effective fitting and is easily solved by Linear Least Squares (LLS), which greatly improves the generalization to unseen concentration of metabolites in the training data. Experimental results show that compared with LCModel, the proposed method has smaller quantification errors for 700 sets of simulated test data, and presents more stable quantification results for 20 sets of healthy in vivo data at a wide range of signal-to-noise ratio. Qnet also outperforms other deep learning methods in terms of lower quantification error on most metabolites. Finally, QNet has been deployed on a cloud computing platform, CloudBrain-MRS, which is open accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge