Taishan Kang
One for Multiple: Physics-informed Synthetic Data Boosts Generalizable Deep Learning for Fast MRI Reconstruction
Jul 25, 2023
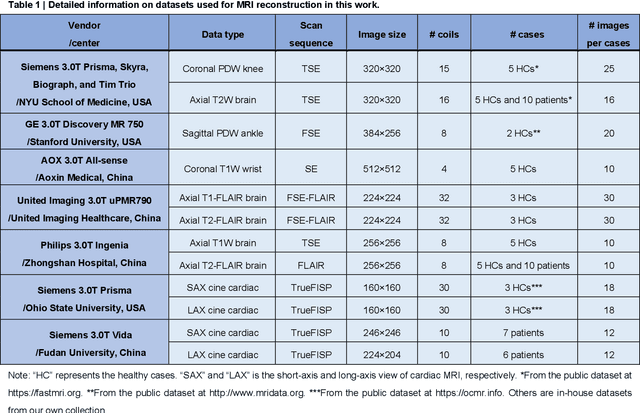
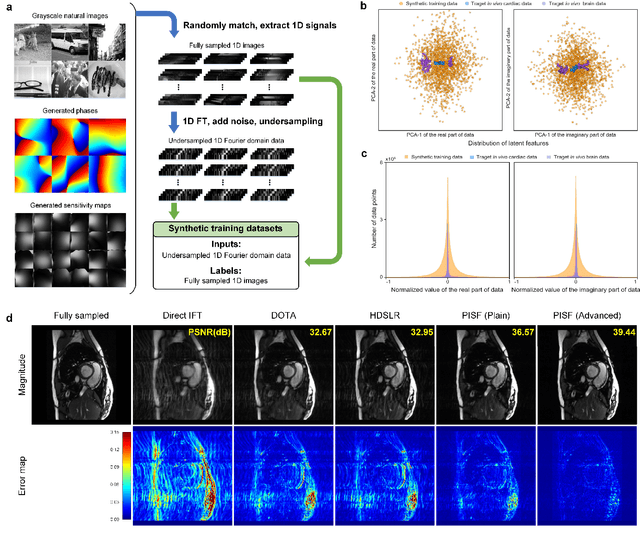
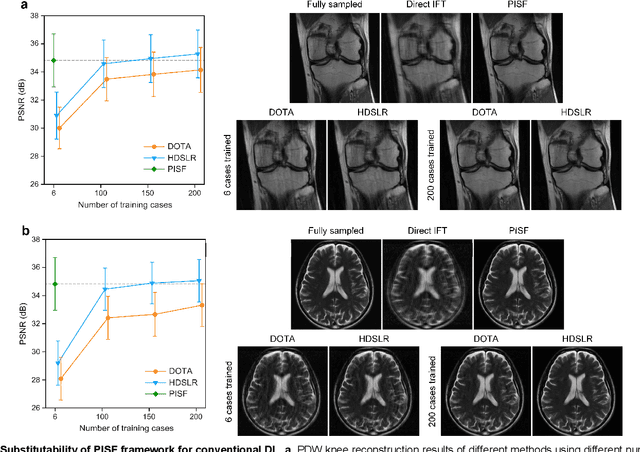
Abstract:Magnetic resonance imaging (MRI) is a principal radiological modality that provides radiation-free, abundant, and diverse information about the whole human body for medical diagnosis, but suffers from prolonged scan time. The scan time can be significantly reduced through k-space undersampling but the introduced artifacts need to be removed in image reconstruction. Although deep learning (DL) has emerged as a powerful tool for image reconstruction in fast MRI, its potential in multiple imaging scenarios remains largely untapped. This is because not only collecting large-scale and diverse realistic training data is generally costly and privacy-restricted, but also existing DL methods are hard to handle the practically inevitable mismatch between training and target data. Here, we present a Physics-Informed Synthetic data learning framework for Fast MRI, called PISF, which is the first to enable generalizable DL for multi-scenario MRI reconstruction using solely one trained model. For a 2D image, the reconstruction is separated into many 1D basic problems and starts with the 1D data synthesis, to facilitate generalization. We demonstrate that training DL models on synthetic data, integrated with enhanced learning techniques, can achieve comparable or even better in vivo MRI reconstruction compared to models trained on a matched realistic dataset, reducing the demand for real-world MRI data by up to 96%. Moreover, our PISF shows impressive generalizability in multi-vendor multi-center imaging. Its excellent adaptability to patients has been verified through 10 experienced doctors' evaluations. PISF provides a feasible and cost-effective way to markedly boost the widespread usage of DL in various fast MRI applications, while freeing from the intractable ethical and practical considerations of in vivo human data acquisitions.
CloudBrain-MRS: An Intelligent Cloud Computing Platform for in vivo Magnetic Resonance Spectroscopy Preprocessing, Quantification, and Analysis
Jun 19, 2023
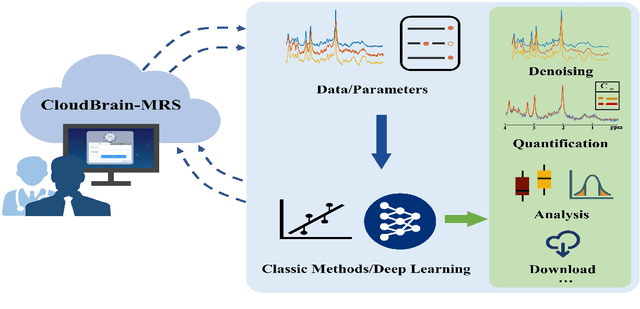
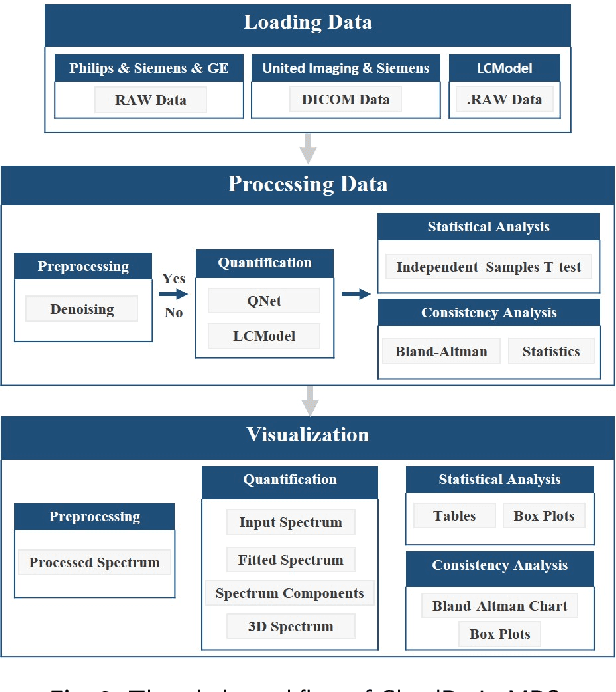
Abstract:Magnetic resonance spectroscopy (MRS) is an important clinical imaging method for the diagnosis of diseases. Spectrum is used to observe the signal intensity of metabolites or further infer their concentrations. Although the magnetic resonance vendors commonly provide basic functions of spectra plots and metabolite quantification, the widespread clinical research of MRS is still limited due to the lack of easy-to-use processing software or platform. To address this issue, we have developed CloudBrain-MRS, a cloud-based online platform that provides powerful hardware and advanced algorithms. The platform can be accessed simply through a web browser, without requiring any program installation on the user side. CloudBrain-MRS also integrates the classic LCModel and advanced artificial intelligence algorithms and supports batch preprocessing, quantification, and analysis of MRS data. Additionally, the platform offers useful functions: 1) Automatically statistical analysis to find biomarkers from the health and patient groups; 2) Consistency verification between the classic and artificial intelligence quantification algorithms; 3) Colorful and three-dimensional visualization for the easy observation of individual metabolite spectrum. Last, both healthy and mild cognitive impairment patient data are used to demonstrate the usefulness of the platform. To the best of our knowledge, this is the first cloud computing platform for in vivo MRS with artificial intelligence processing. We sincerely hope that this platform will facilitate efficient clinical research for MRS. CloudBrain-MRS is open-accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
Magnetic Resonance Spectroscopy Quantification Aided by Deep Estimations of Imperfection Factors and Overall Macromolecular Signal
Jun 16, 2023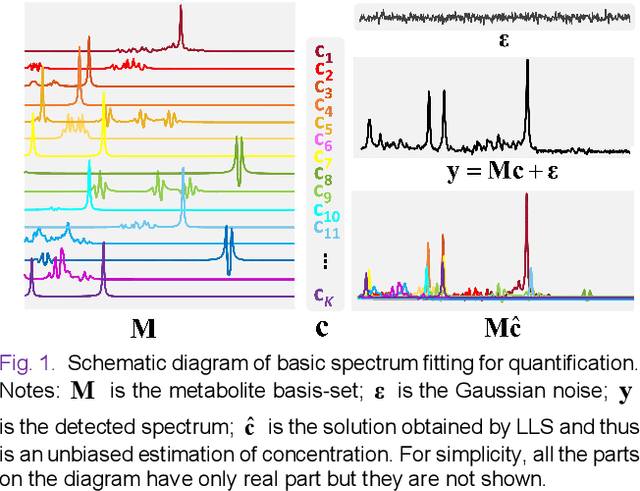
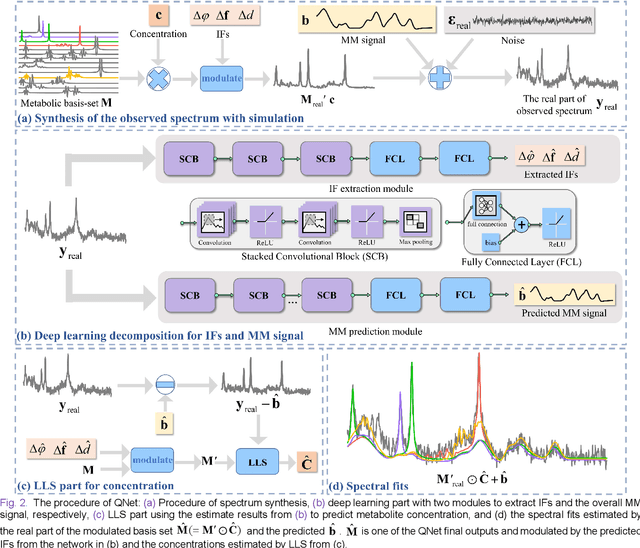
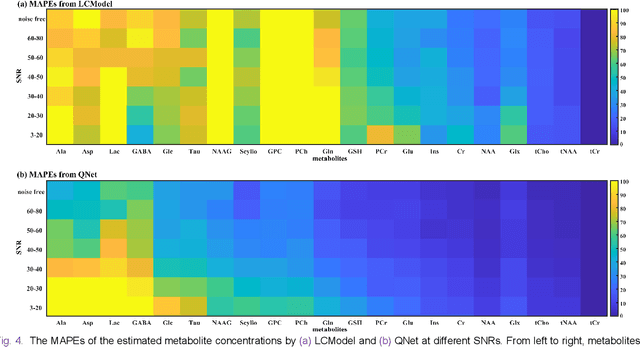
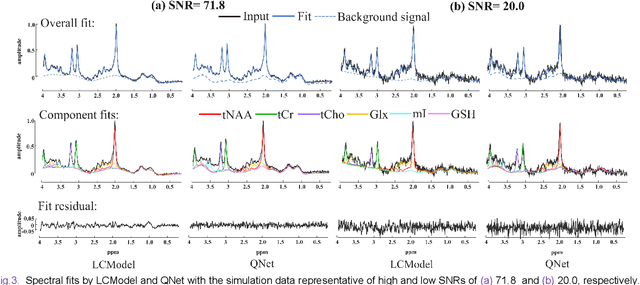
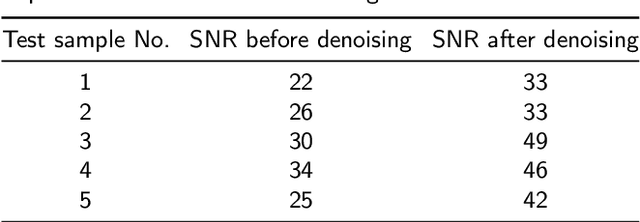
Abstract:Magnetic Resonance Spectroscopy (MRS) is an important non-invasive technique for in vivo biomedical detection. However, it is still challenging to accurately quantify metabolites with proton MRS due to three problems: Serious overlaps of metabolite signals, signal distortions due to non-ideal acquisition conditions and interference with strong background signals including macromolecule signals. The most popular software, LCModel, adopts the non-linear least square to quantify metabolites and addresses these problems by introducing regularization terms, imperfection factors of non-ideal acquisition conditions, and designing several empirical priors such as basissets of both metabolites and macromolecules. However, solving such a large non-linear quantitative problem is complicated. Moreover, when the signal-to-noise ratio of an input MRS signal is low, the solution may have a large deviation. In this work, deep learning is introduced to reduce the complexity of solving this overall quantitative problem. Deep learning is designed to predict directly the imperfection factors and the overall signal from macromolecules. Then, the remaining part of the quantification problem becomes a much simpler effective fitting and is easily solved by Linear Least Squares (LLS), which greatly improves the generalization to unseen concentration of metabolites in the training data. Experimental results show that compared with LCModel, the proposed method has smaller quantification errors for 700 sets of simulated test data, and presents more stable quantification results for 20 sets of healthy in vivo data at a wide range of signal-to-noise ratio. Qnet also outperforms other deep learning methods in terms of lower quantification error on most metabolites. Finally, QNet has been deployed on a cloud computing platform, CloudBrain-MRS, which is open accessed at https://csrc.xmu.edu.cn/CloudBrain.html.
CloudBrain-ReconAI: An Online Platform for MRI Reconstruction and Image Quality Evaluation
Dec 04, 2022



Abstract:Efficient collaboration between engineers and radiologists is important for image reconstruction algorithm development and image quality evaluation in magnetic resonance imaging (MRI). Here, we develop CloudBrain-ReconAI, an online cloud computing platform, for algorithm deployment, fast and blind reader study. This platform supports online image reconstruction using state-of-the-art artificial intelligence and compressed sensing algorithms with applications to fast imaging and high-resolution diffusion imaging. Through visiting the website, radiologists can easily score and mark the images. Then, automatic statistical analysis will be provided. CloudBrain-ReconAI is now open accessed at https://csrc.xmu.edu.cn/CloudBrain.html and will be continually improved to serve the MRI research community.
Physics-informed deep diffusion MRI reconstruction: break the bottleneck of training data in artificial intelligence
Oct 20, 2022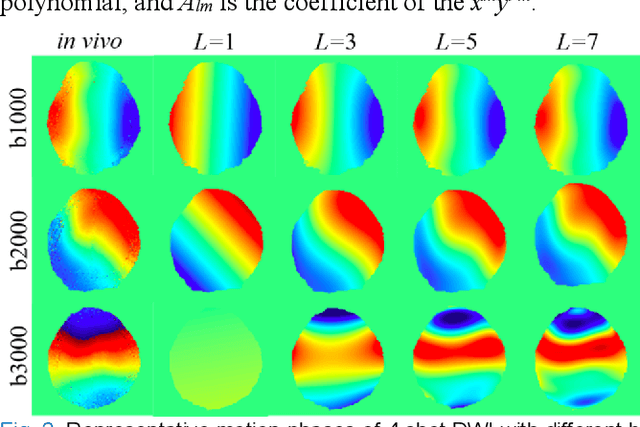
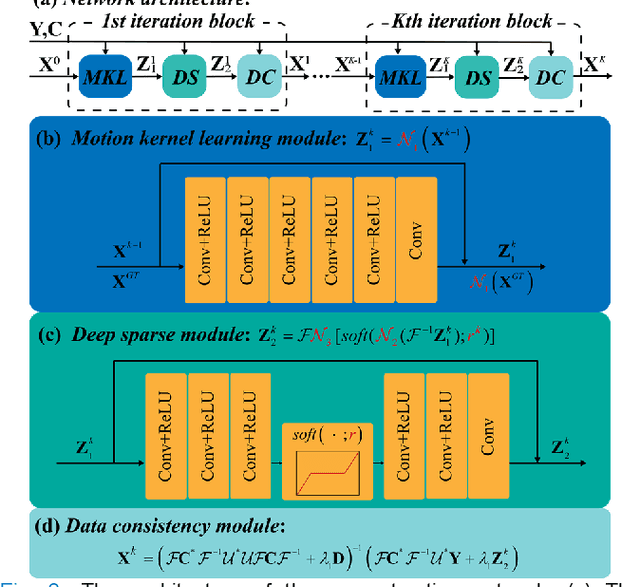
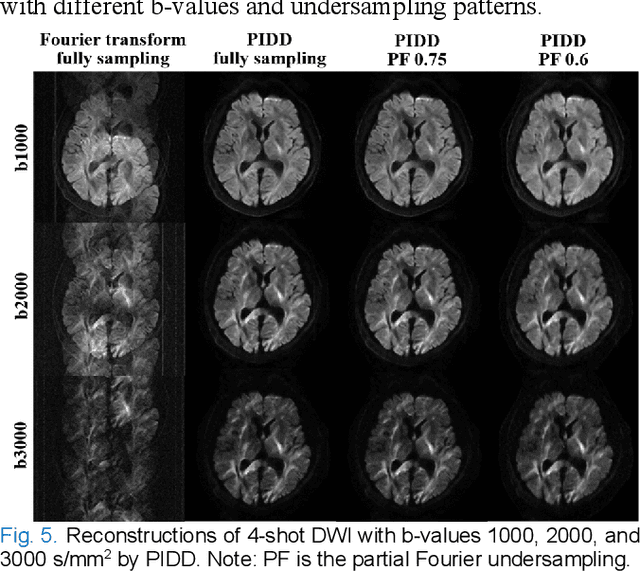
Abstract:In this work, we propose a Physics-Informed Deep Diffusion magnetic resonance imaging (DWI) reconstruction method (PIDD). PIDD contains two main components: The multi-shot DWI data synthesis and a deep learning reconstruction network. For data synthesis, we first mathematically analyze the motion during the multi-shot data acquisition and approach it by a simplified physical motion model. The motion model inspires a polynomial model for motion-induced phase synthesis. Then, lots of synthetic phases are combined with a few real data to generate a large amount of training data. For reconstruction network, we exploit the smoothness property of each shot image phase as learnable convolution kernels in the k-space and complementary sparsity in the image domain. Results on both synthetic and in vivo brain data show that, the proposed PIDD trained on synthetic data enables sub-second ultra-fast, high-quality, and robust reconstruction with different b-values and undersampling patterns.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge