Dongdong Liu
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
Large Scale Unsupervised Brain MRI Image Registration Solution for Learn2Reg 2024
Sep 04, 2024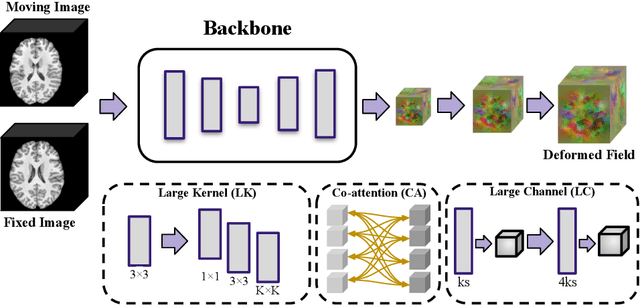
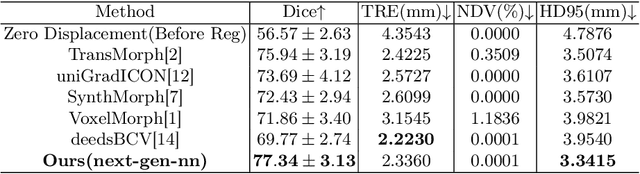
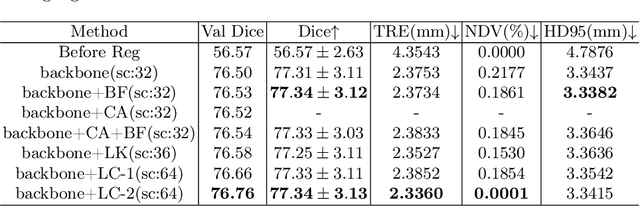
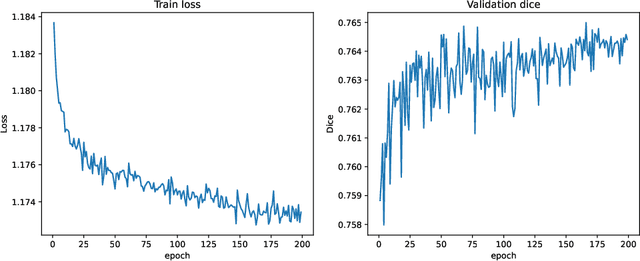
Abstract:In this paper, we summarize the methods and experimental results we proposed for Task 2 in the learn2reg 2024 Challenge. This task focuses on unsupervised registration of anatomical structures in brain MRI images between different patients. The difficulty lies in: (1) without segmentation labels, and (2) a large amount of data. To address these challenges, we built an efficient backbone network and explored several schemes to further enhance registration accuracy. Under the guidance of the NCC loss function and smoothness regularization loss function, we obtained a smooth and reasonable deformation field. According to the leaderboard, our method achieved a Dice coefficient of 77.34%, which is 1.4% higher than the TransMorph. Overall, we won second place on the leaderboard for Task 2.
MemWarp: Discontinuity-Preserving Cardiac Registration with Memorized Anatomical Filters
Jul 10, 2024Abstract:Many existing learning-based deformable image registration methods impose constraints on deformation fields to ensure they are globally smooth and continuous. However, this assumption does not hold in cardiac image registration, where different anatomical regions exhibit asymmetric motions during respiration and movements due to sliding organs within the chest. Consequently, such global constraints fail to accommodate local discontinuities across organ boundaries, potentially resulting in erroneous and unrealistic displacement fields. In this paper, we address this issue with MemWarp, a learning framework that leverages a memory network to store prototypical information tailored to different anatomical regions. MemWarp is different from earlier approaches in two main aspects: firstly, by decoupling feature extraction from similarity matching in moving and fixed images, it facilitates more effective utilization of feature maps; secondly, despite its capability to preserve discontinuities, it eliminates the need for segmentation masks during model inference. In experiments on a publicly available cardiac dataset, our method achieves considerable improvements in registration accuracy and producing realistic deformations, outperforming state-of-the-art methods with a remarkable 7.1\% Dice score improvement over the runner-up semi-supervised method. Source code will be available at https://github.com/tinymilky/Mem-Warp.
Slicer Networks
Jan 18, 2024Abstract:In medical imaging, scans often reveal objects with varied contrasts but consistent internal intensities or textures. This characteristic enables the use of low-frequency approximations for tasks such as segmentation and deformation field estimation. Yet, integrating this concept into neural network architectures for medical image analysis remains underexplored. In this paper, we propose the Slicer Network, a novel architecture designed to leverage these traits. Comprising an encoder utilizing models like vision transformers for feature extraction and a slicer employing a learnable bilateral grid, the Slicer Network strategically refines and upsamples feature maps via a splatting-blurring-slicing process. This introduces an edge-preserving low-frequency approximation for the network outcome, effectively enlarging the effective receptive field. The enhancement not only reduces computational complexity but also boosts overall performance. Experiments across different medical imaging applications, including unsupervised and keypoints-based image registration and lesion segmentation, have verified the Slicer Network's improved accuracy and efficiency.
Spatially Covariant Image Registration with Text Prompts
Nov 27, 2023



Abstract:Medical images are often characterized by their structured anatomical representations and spatially inhomogeneous contrasts. Leveraging anatomical priors in neural networks can greatly enhance their utility in resource-constrained clinical settings. Prior research has harnessed such information for image segmentation, yet progress in deformable image registration has been modest. Our work introduces textSCF, a novel method that integrates spatially covariant filters and textual anatomical prompts encoded by visual-language models, to fill this gap. This approach optimizes an implicit function that correlates text embeddings of anatomical regions to filter weights, relaxing the typical translation-invariance constraint of convolutional operations. TextSCF not only boosts computational efficiency but can also retain or improve registration accuracy. By capturing the contextual interplay between anatomical regions, it offers impressive inter-regional transferability and the ability to preserve structural discontinuities during registration. TextSCF's performance has been rigorously tested on inter-subject brain MRI and abdominal CT registration tasks, outperforming existing state-of-the-art models in the MICCAI Learn2Reg 2021 challenge and leading the leaderboard. In abdominal registrations, textSCF's larger model variant improved the Dice score by 11.3% over the second-best model, while its smaller variant maintained similar accuracy but with an 89.13% reduction in network parameters and a 98.34\% decrease in computational operations.
Spatially Covariant Lesion Segmentation
Jan 19, 2023



Abstract:Compared to natural images, medical images usually show stronger visual patterns and therefore this adds flexibility and elasticity to resource-limited clinical applications by injecting proper priors into neural networks. In this paper, we propose spatially covariant pixel-aligned classifier (SCP) to improve the computational efficiency and meantime maintain or increase accuracy for lesion segmentation. SCP relaxes the spatial invariance constraint imposed by convolutional operations and optimizes an underlying implicit function that maps image coordinates to network weights, the parameters of which are obtained along with the backbone network training and later used for generating network weights to capture spatially covariant contextual information. We demonstrate the effectiveness and efficiency of the proposed SCP using two lesion segmentation tasks from different imaging modalities: white matter hyperintensity segmentation in magnetic resonance imaging and liver tumor segmentation in contrast-enhanced abdominal computerized tomography. The network using SCP has achieved 23.8%, 64.9% and 74.7% reduction in GPU memory usage, FLOPs, and network size with similar or better accuracy for lesion segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge