Mathilde Antoniades
Dimensional Neuroimaging Endophenotypes: Neurobiological Representations of Disease Heterogeneity Through Machine Learning
Jan 17, 2024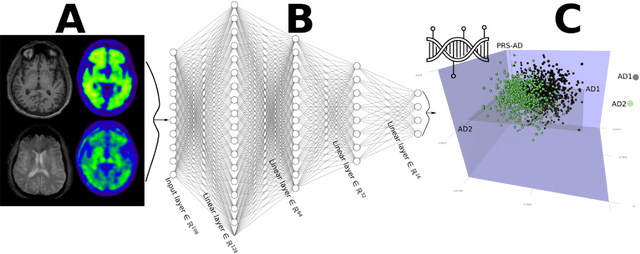
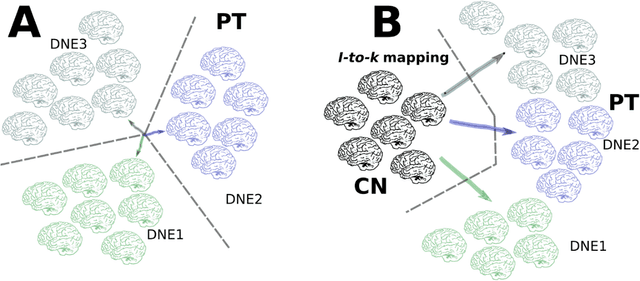
Abstract:Machine learning has been increasingly used to obtain individualized neuroimaging signatures for disease diagnosis, prognosis, and response to treatment in neuropsychiatric and neurodegenerative disorders. Therefore, it has contributed to a better understanding of disease heterogeneity by identifying disease subtypes that present significant differences in various brain phenotypic measures. In this review, we first present a systematic literature overview of studies using machine learning and multimodal MRI to unravel disease heterogeneity in various neuropsychiatric and neurodegenerative disorders, including Alzheimer disease, schizophrenia, major depressive disorder, autism spectrum disorder, multiple sclerosis, as well as their potential in transdiagnostic settings. Subsequently, we summarize relevant machine learning methodologies and discuss an emerging paradigm which we call dimensional neuroimaging endophenotype (DNE). DNE dissects the neurobiological heterogeneity of neuropsychiatric and neurodegenerative disorders into a low dimensional yet informative, quantitative brain phenotypic representation, serving as a robust intermediate phenotype (i.e., endophenotype) largely reflecting underlying genetics and etiology. Finally, we discuss the potential clinical implications of the current findings and envision future research avenues.
Robust Hierarchical Patterns for identifying MDD patients: A Multisite Study
Feb 22, 2022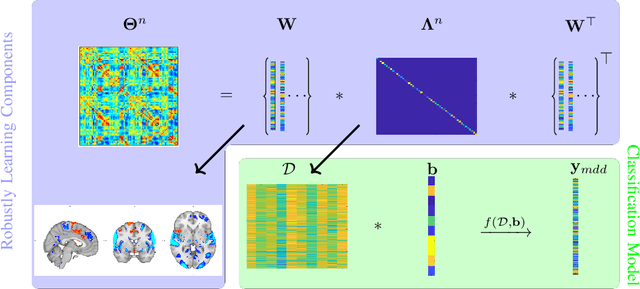
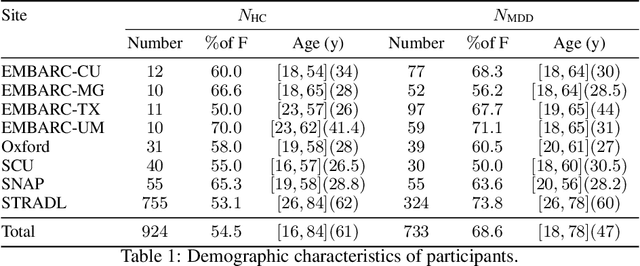
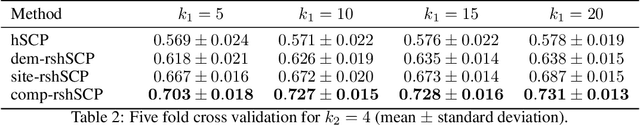
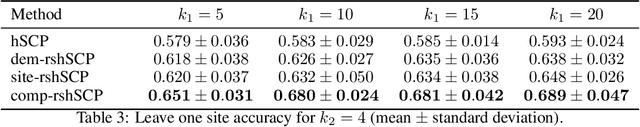
Abstract:Many supervised machine learning frameworks have been proposed for disease classification using functional magnetic resonance imaging (fMRI) data, producing important biomarkers. More recently, data pooling has flourished, making the result generalizable across a large population. But, this success depends on the population diversity and variability introduced due to the pooling of the data that is not a primary research interest. Here, we look at hierarchical Sparse Connectivity Patterns (hSCPs) as biomarkers for major depressive disorder (MDD). We propose a novel model based on hSCPs to predict MDD patients from functional connectivity matrices extracted from resting-state fMRI data. Our model consists of three coupled terms. The first term decomposes connectivity matrices into hierarchical low-rank sparse components corresponding to synchronous patterns across the human brain. These components are then combined via patient-specific weights capturing heterogeneity in the data. The second term is a classification loss that uses the patient-specific weights to classify MDD patients from healthy ones. Both of these terms are combined with the third term, a robustness loss function to improve the reproducibility of hSCPs. This reduces the variability introduced due to site and population diversity (age and sex) on the predictive accuracy and pattern stability in a large dataset pooled from five different sites. Our results show the impact of diversity on prediction performance. Our model can reduce diversity and improve the predictive and generalizing capability of the components. Finally, our results show that our proposed model can robustly identify clinically relevant patterns characteristic of MDD with high reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge