Ioanna Skampardoni
A comprehensive interpretable machine learning framework for Mild Cognitive Impairment and Alzheimer's disease diagnosis
Dec 12, 2024



Abstract:An interpretable machine learning (ML) framework is introduced to enhance the diagnosis of Mild Cognitive Impairment (MCI) and Alzheimer's disease (AD) by ensuring robustness of the ML models' interpretations. The dataset used comprises volumetric measurements from brain MRI and genetic data from healthy individuals and patients with MCI/AD, obtained through the Alzheimer's Disease Neuroimaging Initiative. The existing class imbalance is addressed by an ensemble learning approach, while various attribution-based and counterfactual-based interpretability methods are leveraged towards producing diverse explanations related to the pathophysiology of MCI/AD. A unification method combining SHAP with counterfactual explanations assesses the interpretability techniques' robustness. The best performing model yielded 87.5% balanced accuracy and 90.8% F1-score. The attribution-based interpretability methods highlighted significant volumetric and genetic features related to MCI/AD risk. The unification method provided useful insights regarding those features' necessity and sufficiency, further showcasing their significance in MCI/AD diagnosis.
Dimensional Neuroimaging Endophenotypes: Neurobiological Representations of Disease Heterogeneity Through Machine Learning
Jan 17, 2024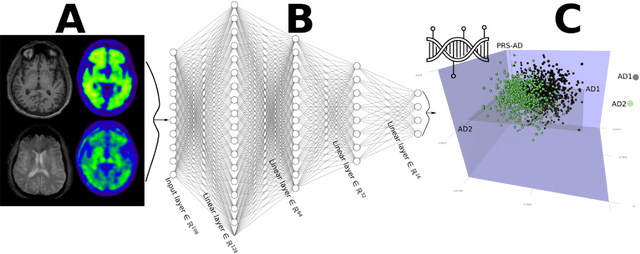
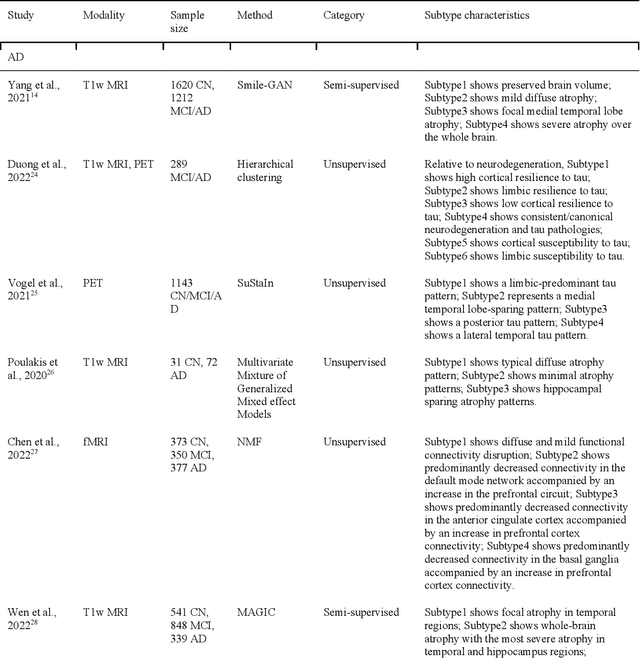
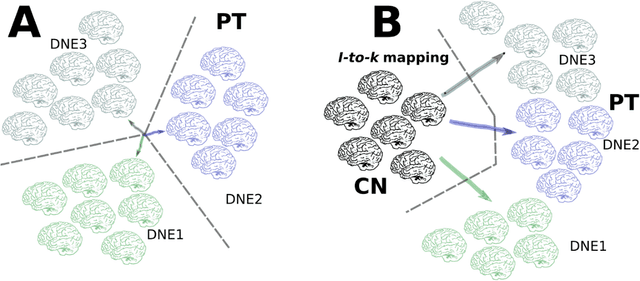
Abstract:Machine learning has been increasingly used to obtain individualized neuroimaging signatures for disease diagnosis, prognosis, and response to treatment in neuropsychiatric and neurodegenerative disorders. Therefore, it has contributed to a better understanding of disease heterogeneity by identifying disease subtypes that present significant differences in various brain phenotypic measures. In this review, we first present a systematic literature overview of studies using machine learning and multimodal MRI to unravel disease heterogeneity in various neuropsychiatric and neurodegenerative disorders, including Alzheimer disease, schizophrenia, major depressive disorder, autism spectrum disorder, multiple sclerosis, as well as their potential in transdiagnostic settings. Subsequently, we summarize relevant machine learning methodologies and discuss an emerging paradigm which we call dimensional neuroimaging endophenotype (DNE). DNE dissects the neurobiological heterogeneity of neuropsychiatric and neurodegenerative disorders into a low dimensional yet informative, quantitative brain phenotypic representation, serving as a robust intermediate phenotype (i.e., endophenotype) largely reflecting underlying genetics and etiology. Finally, we discuss the potential clinical implications of the current findings and envision future research avenues.
Applications of Generative Adversarial Networks in Neuroimaging and Clinical Neuroscience
Jun 14, 2022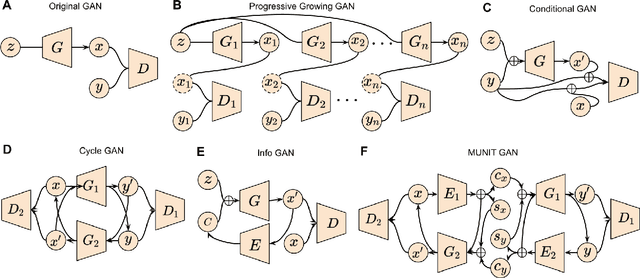
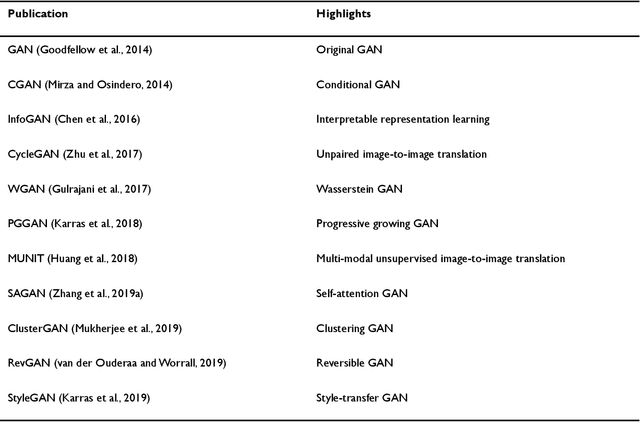
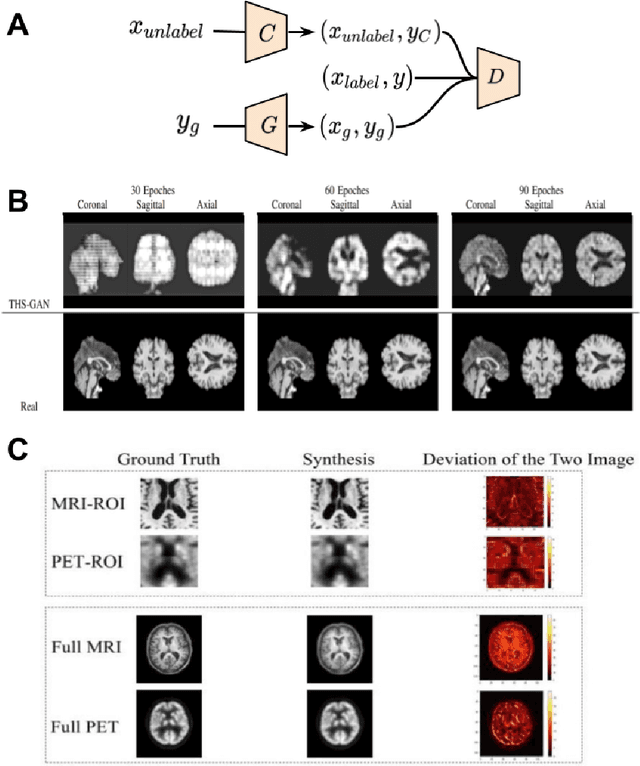
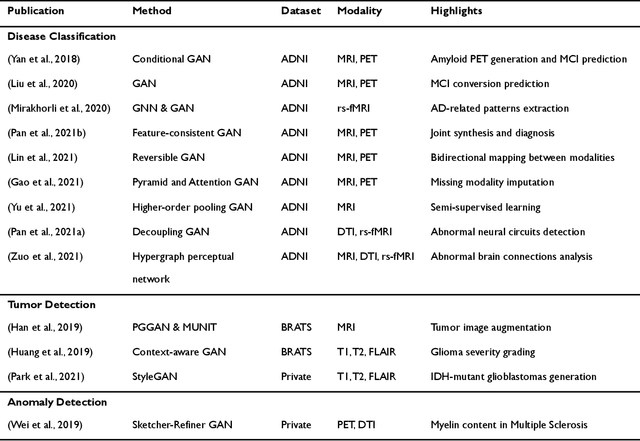
Abstract:Generative adversarial networks (GANs) are one powerful type of deep learning models that have been successfully utilized in numerous fields. They belong to a broader family called generative methods, which generate new data with a probabilistic model by learning sample distribution from real examples. In the clinical context, GANs have shown enhanced capabilities in capturing spatially complex, nonlinear, and potentially subtle disease effects compared to traditional generative methods. This review appraises the existing literature on the applications of GANs in imaging studies of various neurological conditions, including Alzheimer's disease, brain tumors, brain aging, and multiple sclerosis. We provide an intuitive explanation of various GAN methods for each application and further discuss the main challenges, open questions, and promising future directions of leveraging GANs in neuroimaging. We aim to bridge the gap between advanced deep learning methods and neurology research by highlighting how GANs can be leveraged to support clinical decision making and contribute to a better understanding of the structural and functional patterns of brain diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge