Vasiliki Tassopoulou
Uncertainty-Calibrated Prediction of Randomly-Timed Biomarker Trajectories with Conformal Bands
Nov 17, 2025Abstract:Despite recent progress in predicting biomarker trajectories from real clinical data, uncertainty in the predictions poses high-stakes risks (e.g., misdiagnosis) that limit their clinical deployment. To enable safe and reliable use of such predictions in healthcare, we introduce a conformal method for uncertainty-calibrated prediction of biomarker trajectories resulting from randomly-timed clinical visits of patients. Our approach extends conformal prediction to the setting of randomly-timed trajectories via a novel nonconformity score that produces prediction bands guaranteed to cover the unknown biomarker trajectories with a user-prescribed probability. We apply our method across a wide range of standard and state-of-the-art predictors for two well-established brain biomarkers of Alzheimer's disease, using neuroimaging data from real clinical studies. We observe that our conformal prediction bands consistently achieve the desired coverage, while also being tighter than baseline prediction bands. To further account for population heterogeneity, we develop group-conditional conformal bands and test their coverage guarantees across various demographic and clinically relevant subpopulations. Moreover, we demonstrate the clinical utility of our conformal bands in identifying subjects at high risk of progression to Alzheimer's disease. Specifically, we introduce an uncertainty-calibrated risk score that enables the identification of 17.5% more high-risk subjects compared to standard risk scores, highlighting the value of uncertainty calibration in real-world clinical decision making. Our code is available at github.com/vatass/ConformalBiomarkerTrajectories.
Adaptive Shrinkage Estimation For Personalized Deep Kernel Regression In Modeling Brain Trajectories
Apr 10, 2025



Abstract:Longitudinal biomedical studies monitor individuals over time to capture dynamics in brain development, disease progression, and treatment effects. However, estimating trajectories of brain biomarkers is challenging due to biological variability, inconsistencies in measurement protocols (e.g., differences in MRI scanners), scarcity, and irregularity in longitudinal measurements. Herein, we introduce a novel personalized deep kernel regression framework for forecasting brain biomarkers, with application to regional volumetric measurements. Our approach integrates two key components: a population model that captures brain trajectories from a large and diverse cohort, and a subject-specific model that captures individual trajectories. To optimally combine these, we propose Adaptive Shrinkage Estimation, which effectively balances population and subject-specific models. We assess our model's performance through predictive accuracy metrics, uncertainty quantification, and validation against external clinical studies. Benchmarking against state-of-the-art statistical and machine learning models -- including linear mixed effects models, generalized additive models, and deep learning methods -- demonstrates the superior predictive performance of our approach. Additionally, we apply our method to predict trajectories of composite neuroimaging biomarkers, which highlights the versatility of our approach in modeling the progression of longitudinal neuroimaging biomarkers. Furthermore, validation on three external neuroimaging studies confirms the robustness of our method across different clinical contexts. We make the code available at https://github.com/vatass/AdaptiveShrinkageDKGP.
NeuroSynth: MRI-Derived Neuroanatomical Generative Models and Associated Dataset of 18,000 Samples
Jul 17, 2024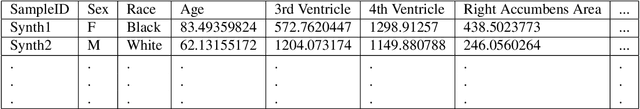
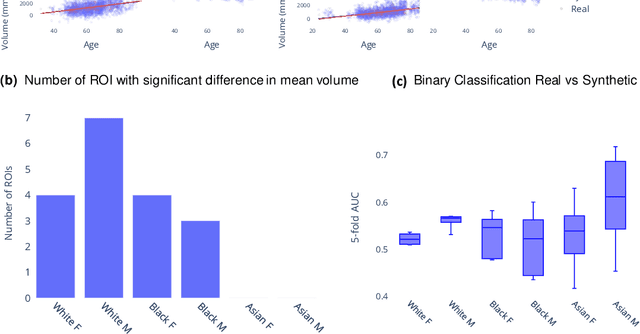
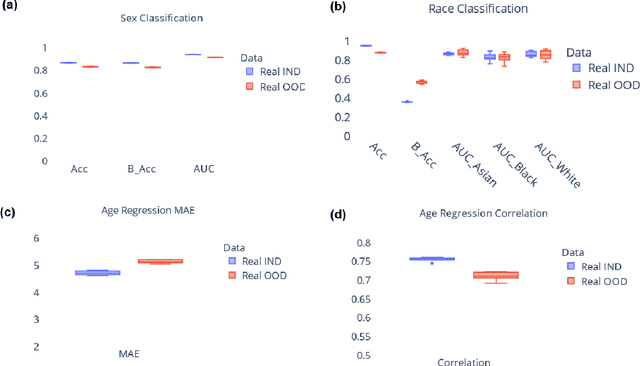
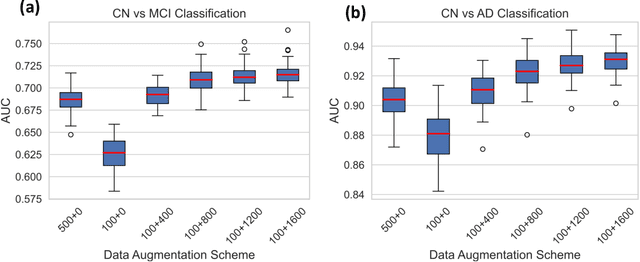
Abstract:Availability of large and diverse medical datasets is often challenged by privacy and data sharing restrictions. For successful application of machine learning techniques for disease diagnosis, prognosis, and precision medicine, large amounts of data are necessary for model building and optimization. To help overcome such limitations in the context of brain MRI, we present NeuroSynth: a collection of generative models of normative regional volumetric features derived from structural brain imaging. NeuroSynth models are trained on real brain imaging regional volumetric measures from the iSTAGING consortium, which encompasses over 40,000 MRI scans across 13 studies, incorporating covariates such as age, sex, and race. Leveraging NeuroSynth, we produce and offer 18,000 synthetic samples spanning the adult lifespan (ages 22-90 years), alongside the model's capability to generate unlimited data. Experimental results indicate that samples generated from NeuroSynth agree with the distributions obtained from real data. Most importantly, the generated normative data significantly enhance the accuracy of downstream machine learning models on tasks such as disease classification. Data and models are available at: https://huggingface.co/spaces/rongguangw/neuro-synth.
Applications of Generative Adversarial Networks in Neuroimaging and Clinical Neuroscience
Jun 14, 2022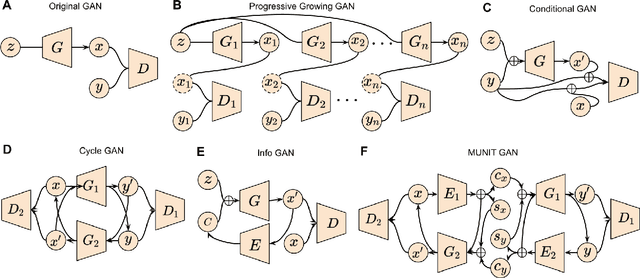
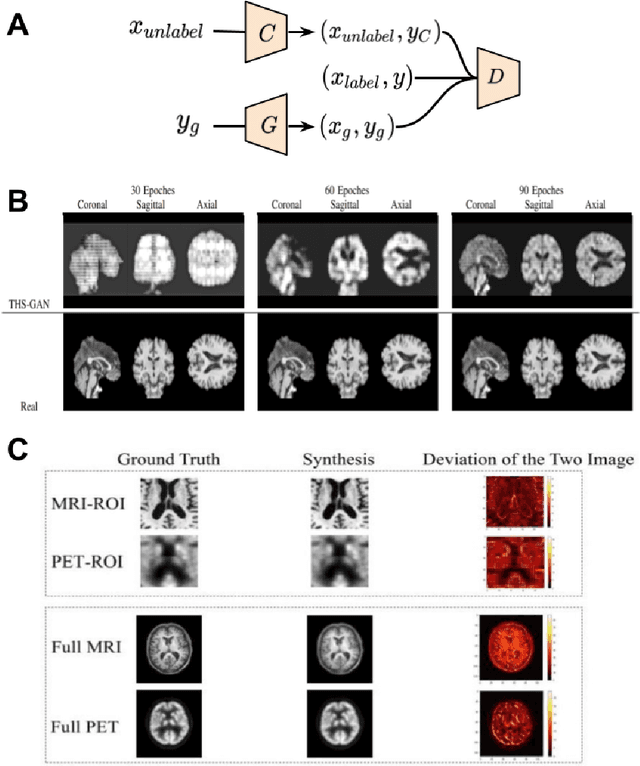
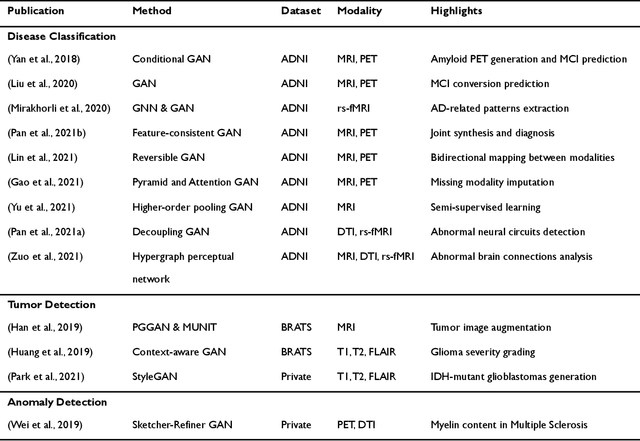
Abstract:Generative adversarial networks (GANs) are one powerful type of deep learning models that have been successfully utilized in numerous fields. They belong to a broader family called generative methods, which generate new data with a probabilistic model by learning sample distribution from real examples. In the clinical context, GANs have shown enhanced capabilities in capturing spatially complex, nonlinear, and potentially subtle disease effects compared to traditional generative methods. This review appraises the existing literature on the applications of GANs in imaging studies of various neurological conditions, including Alzheimer's disease, brain tumors, brain aging, and multiple sclerosis. We provide an intuitive explanation of various GAN methods for each application and further discuss the main challenges, open questions, and promising future directions of leveraging GANs in neuroimaging. We aim to bridge the gap between advanced deep learning methods and neurology research by highlighting how GANs can be leveraged to support clinical decision making and contribute to a better understanding of the structural and functional patterns of brain diseases.
Enhancing Handwritten Text Recognition with N-gram sequence decomposition and Multitask Learning
Dec 28, 2020
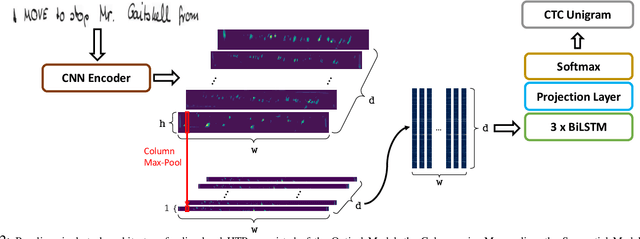
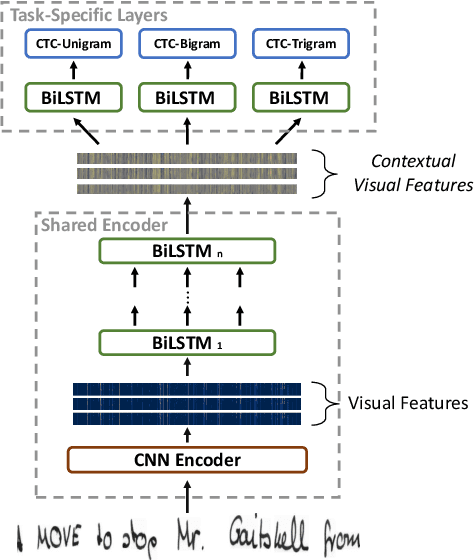
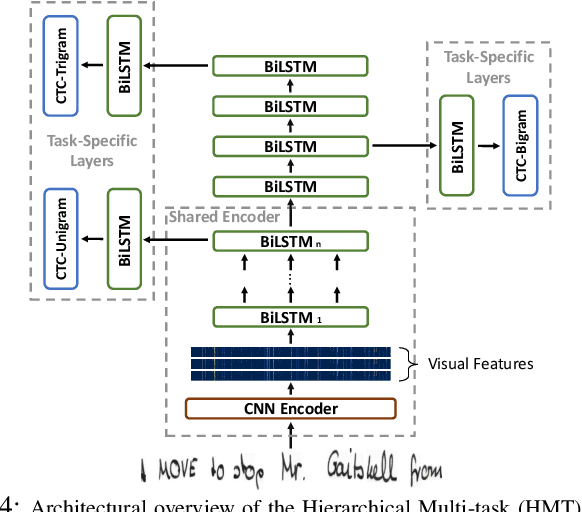
Abstract:Current state-of-the-art approaches in the field of Handwritten Text Recognition are predominately single task with unigram, character level target units. In our work, we utilize a Multi-task Learning scheme, training the model to perform decompositions of the target sequence with target units of different granularity, from fine to coarse. We consider this method as a way to utilize n-gram information, implicitly, in the training process, while the final recognition is performed using only the unigram output. % in order to highlight the difference of the internal Unigram decoding of such a multi-task approach highlights the capability of the learned internal representations, imposed by the different n-grams at the training step. We select n-grams as our target units and we experiment from unigrams to fourgrams, namely subword level granularities. These multiple decompositions are learned from the network with task-specific CTC losses. Concerning network architectures, we propose two alternatives, namely the Hierarchical and the Block Multi-task. Overall, our proposed model, even though evaluated only on the unigram task, outperforms its counterpart single-task by absolute 2.52\% WER and 1.02\% CER, in the greedy decoding, without any computational overhead during inference, hinting towards successfully imposing an implicit language model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge