Dushyant Sahoo
Applications of Generative Adversarial Networks in Neuroimaging and Clinical Neuroscience
Jun 14, 2022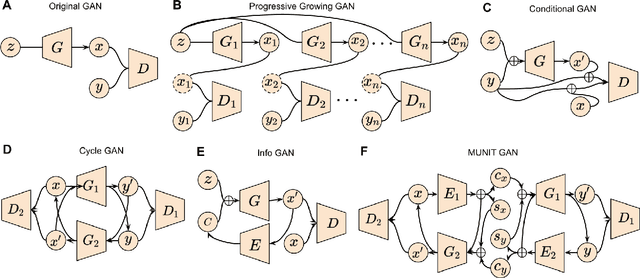
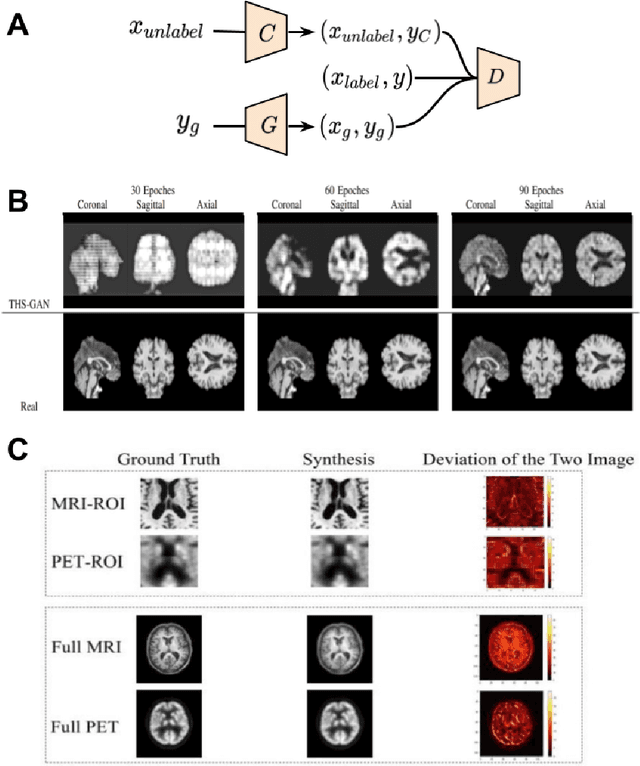
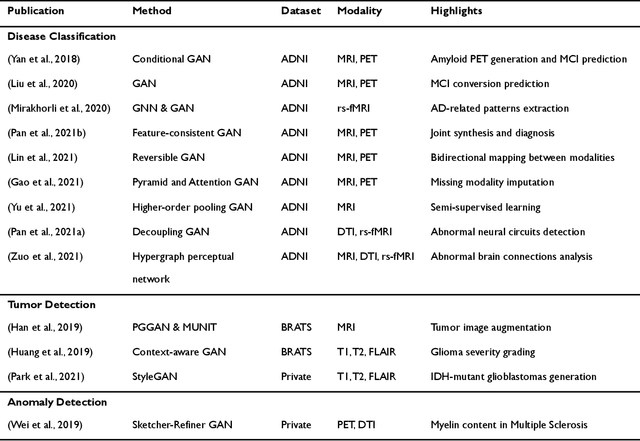
Abstract:Generative adversarial networks (GANs) are one powerful type of deep learning models that have been successfully utilized in numerous fields. They belong to a broader family called generative methods, which generate new data with a probabilistic model by learning sample distribution from real examples. In the clinical context, GANs have shown enhanced capabilities in capturing spatially complex, nonlinear, and potentially subtle disease effects compared to traditional generative methods. This review appraises the existing literature on the applications of GANs in imaging studies of various neurological conditions, including Alzheimer's disease, brain tumors, brain aging, and multiple sclerosis. We provide an intuitive explanation of various GAN methods for each application and further discuss the main challenges, open questions, and promising future directions of leveraging GANs in neuroimaging. We aim to bridge the gap between advanced deep learning methods and neurology research by highlighting how GANs can be leveraged to support clinical decision making and contribute to a better understanding of the structural and functional patterns of brain diseases.
Robust Hierarchical Patterns for identifying MDD patients: A Multisite Study
Feb 22, 2022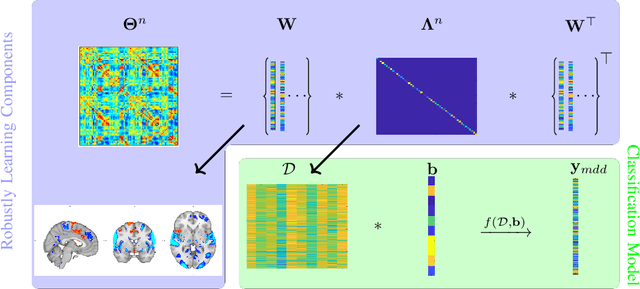
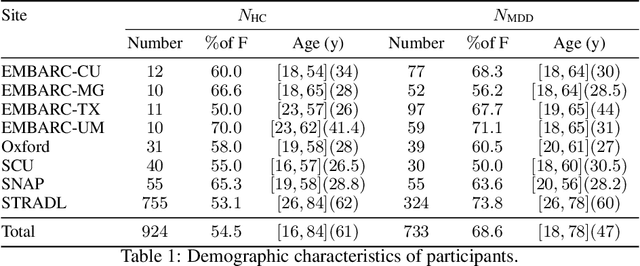
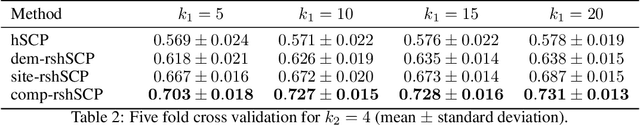
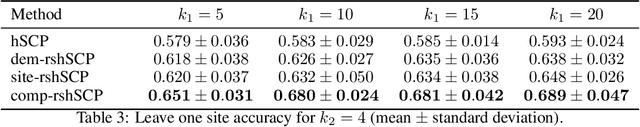
Abstract:Many supervised machine learning frameworks have been proposed for disease classification using functional magnetic resonance imaging (fMRI) data, producing important biomarkers. More recently, data pooling has flourished, making the result generalizable across a large population. But, this success depends on the population diversity and variability introduced due to the pooling of the data that is not a primary research interest. Here, we look at hierarchical Sparse Connectivity Patterns (hSCPs) as biomarkers for major depressive disorder (MDD). We propose a novel model based on hSCPs to predict MDD patients from functional connectivity matrices extracted from resting-state fMRI data. Our model consists of three coupled terms. The first term decomposes connectivity matrices into hierarchical low-rank sparse components corresponding to synchronous patterns across the human brain. These components are then combined via patient-specific weights capturing heterogeneity in the data. The second term is a classification loss that uses the patient-specific weights to classify MDD patients from healthy ones. Both of these terms are combined with the third term, a robustness loss function to improve the reproducibility of hSCPs. This reduces the variability introduced due to site and population diversity (age and sex) on the predictive accuracy and pattern stability in a large dataset pooled from five different sites. Our results show the impact of diversity on prediction performance. Our model can reduce diversity and improve the predictive and generalizing capability of the components. Finally, our results show that our proposed model can robustly identify clinically relevant patterns characteristic of MDD with high reproducibility.
Learning Robust Hierarchical Patterns of Human Brain across Many fMRI Studies
May 13, 2021



Abstract:Resting-state fMRI has been shown to provide surrogate biomarkers for the analysis of various diseases. In addition, fMRI data helps in understanding the brain's functional working during resting state and task-induced activity. To improve the statistical power of biomarkers and the understanding mechanism of the brain, pooling of multi-center studies has become increasingly popular. But pooling the data from multiple sites introduces variations due to hardware, software, and environment. In this paper, we look at the estimation problem of hierarchical Sparsity Connectivity Patterns (hSCPs) in fMRI data acquired on multiple sites. We introduce a simple yet effective matrix factorization based formulation to reduce site-related effects while preserving biologically relevant variations. We leverage adversarial learning in the unsupervised regime to improve the reproducibility of the components. Experiments on simulated datasets display that the proposed method can estimate components with improved accuracy and reproducibility. We also demonstrate the improved reproducibility of the components while preserving age-related variation on a real dataset compiled from multiple sites.
Extraction of Hierarchical Functional Connectivity Components in human brain using Adversarial Learning
Apr 20, 2021



Abstract:The estimation of sparse hierarchical components reflecting patterns of the brain's functional connectivity from rsfMRI data can contribute to our understanding of the brain's functional organization, and can lead to biomarkers of diseases. However, inter-scanner variations and other confounding factors pose a challenge to the robust and reproducible estimation of functionally-interpretable brain networks, and especially to reproducible biomarkers. Moreover, the brain is believed to be organized hierarchically, and hence single-scale decompositions miss this hierarchy. The paper aims to use current advancements in adversarial learning to estimate interpretable hierarchical patterns in the human brain using rsfMRI data, which are robust to "adversarial effects" such as inter-scanner variations. We write the estimation problem as a minimization problem and solve it using alternating updates. Extensive experiments on simulation and a real-world dataset show high reproducibility of the components compared to other well-known methods.
Variance Reduced Stochastic Proximal Algorithm for AUC Maximization
Nov 08, 2019


Abstract:Stochastic Gradient Descent has been widely studied with classification accuracy as a performance measure. However, these stochastic algorithms cannot be directly used when non-decomposable pairwise performance measures are used such as Area under the ROC curve (AUC) which is a common performance metric when the classes are imbalanced. There have been several algorithms proposed for optimizing AUC as a performance metric, and one of the recent being a stochastic proximal gradient algorithm (SPAM). But the downside of the stochastic methods is that they suffer from high variance leading to slower convergence. To combat this issue, several variance reduced methods have been proposed with faster convergence guarantees than vanilla stochastic gradient descent. Again, these variance reduced methods are not directly applicable when non-decomposable performance measures are used. In this paper, we develop a Variance Reduced Stochastic Proximal algorithm for AUC Maximization (\textsc{VRSPAM}) and perform a theoretical analysis as well as empirical analysis to show that our algorithm converges faster than SPAM which is the previous state-of-the-art for the AUC maximization problem.
Extraction of hierarchical functional connectivity components in human brain using resting-state fMRI
Jun 27, 2019



Abstract:The study of hierarchy in networks of the human brain has been of significant interest among the researchers as numerous studies have pointed out towards a functional hierarchical organization of the human brain. This paper provides a novel method for the extraction of hierarchical connectivity components in the human brain using resting-state fMRI. The method builds upon prior work of Sparse Connectivity Patterns (SCPs) by introducing a hierarchy of sparse overlapping patterns. The components are estimated by deep factorization of correlation matrices generated from fMRI. The goal of the paper is to extract interpretable hierarchical patterns using correlation matrices where a low rank decomposition is formed by a linear combination of a high rank decomposition. We formulate the decomposition as a non-convex optimization problem and solve it using gradient descent algorithms with adaptive step size. We also provide a method for the warm start of the gradient descent using singular value decomposition. We demonstrate the effectiveness of the developed method on two different real-world datasets by showing that multi-scale hierarchical SCPs are reproducible between sub-samples and are more reproducible as compared to single scale patterns. We also compare our method with existing hierarchical community detection approaches. Our method also provides novel insight into the functional organization of the human brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge