Jingxuan Bao
Flex-MoE: Modeling Arbitrary Modality Combination via the Flexible Mixture-of-Experts
Oct 10, 2024Abstract:Multimodal learning has gained increasing importance across various fields, offering the ability to integrate data from diverse sources such as images, text, and personalized records, which are frequently observed in medical domains. However, in scenarios where some modalities are missing, many existing frameworks struggle to accommodate arbitrary modality combinations, often relying heavily on a single modality or complete data. This oversight of potential modality combinations limits their applicability in real-world situations. To address this challenge, we propose Flex-MoE (Flexible Mixture-of-Experts), a new framework designed to flexibly incorporate arbitrary modality combinations while maintaining robustness to missing data. The core idea of Flex-MoE is to first address missing modalities using a new missing modality bank that integrates observed modality combinations with the corresponding missing ones. This is followed by a uniquely designed Sparse MoE framework. Specifically, Flex-MoE first trains experts using samples with all modalities to inject generalized knowledge through the generalized router ($\mathcal{G}$-Router). The $\mathcal{S}$-Router then specializes in handling fewer modality combinations by assigning the top-1 gate to the expert corresponding to the observed modality combination. We evaluate Flex-MoE on the ADNI dataset, which encompasses four modalities in the Alzheimer's Disease domain, as well as on the MIMIC-IV dataset. The results demonstrate the effectiveness of Flex-MoE highlighting its ability to model arbitrary modality combinations in diverse missing modality scenarios. Code is available at https://github.com/UNITES-Lab/flex-moe.
Clustering Alzheimer's Disease Subtypes via Similarity Learning and Graph Diffusion
Oct 04, 2024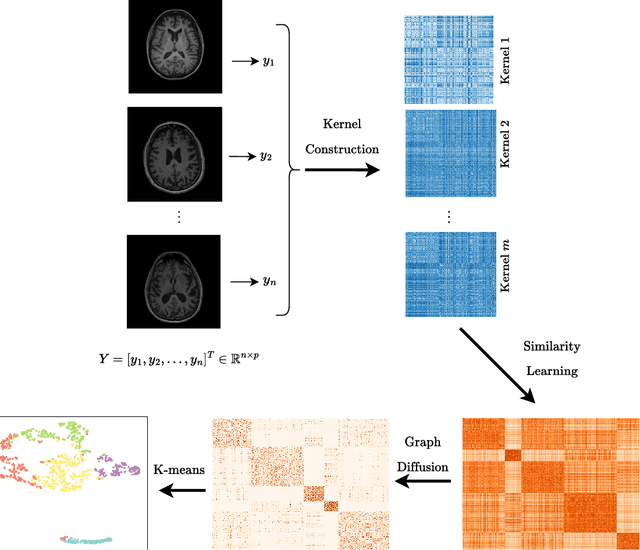
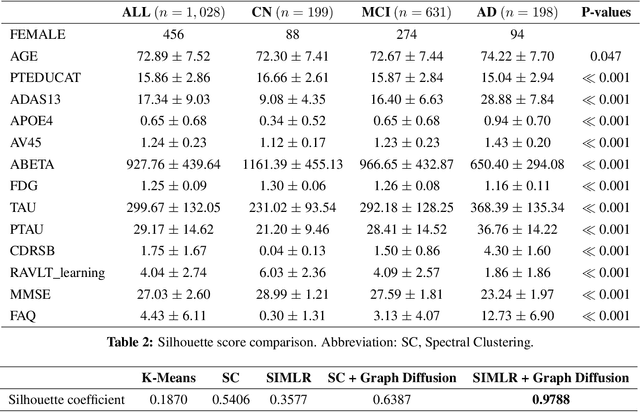
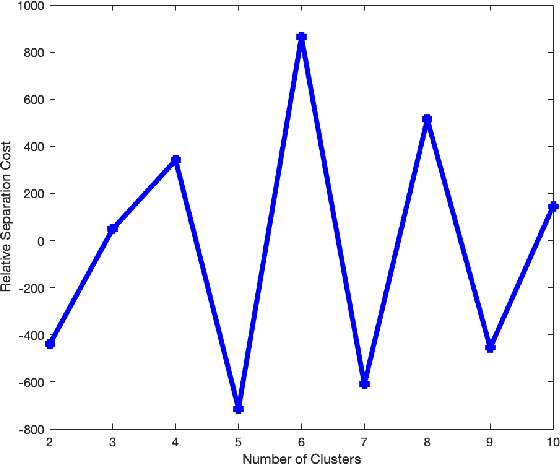
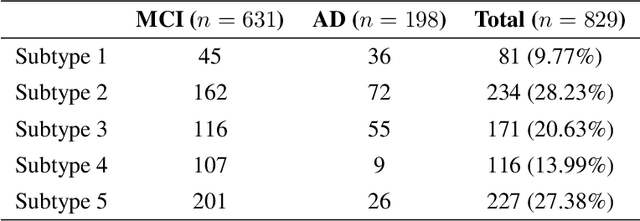
Abstract:Alzheimer's disease (AD) is a complex neurodegenerative disorder that affects millions of people worldwide. Due to the heterogeneous nature of AD, its diagnosis and treatment pose critical challenges. Consequently, there is a growing research interest in identifying homogeneous AD subtypes that can assist in addressing these challenges in recent years. In this study, we aim to identify subtypes of AD that represent distinctive clinical features and underlying pathology by utilizing unsupervised clustering with graph diffusion and similarity learning. We adopted SIMLR, a multi-kernel similarity learning framework, and graph diffusion to perform clustering on a group of 829 patients with AD and mild cognitive impairment (MCI, a prodromal stage of AD) based on their cortical thickness measurements extracted from magnetic resonance imaging (MRI) scans. Although the clustering approach we utilized has not been explored for the task of AD subtyping before, it demonstrated significantly better performance than several commonly used clustering methods. Specifically, we showed the power of graph diffusion in reducing the effects of noise in the subtype detection. Our results revealed five subtypes that differed remarkably in their biomarkers, cognitive status, and some other clinical features. To evaluate the resultant subtypes further, a genetic association study was carried out and successfully identified potential genetic underpinnings of different AD subtypes. Our source code is available at: https://github.com/PennShenLab/AD-SIMLR.
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023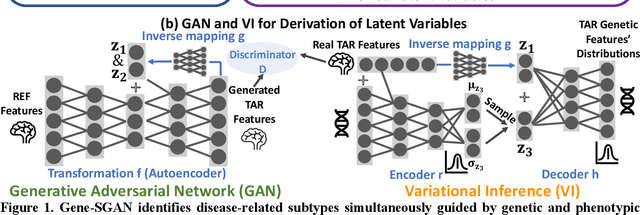
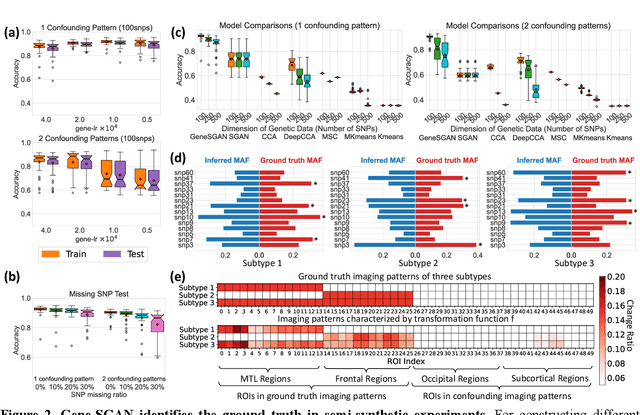
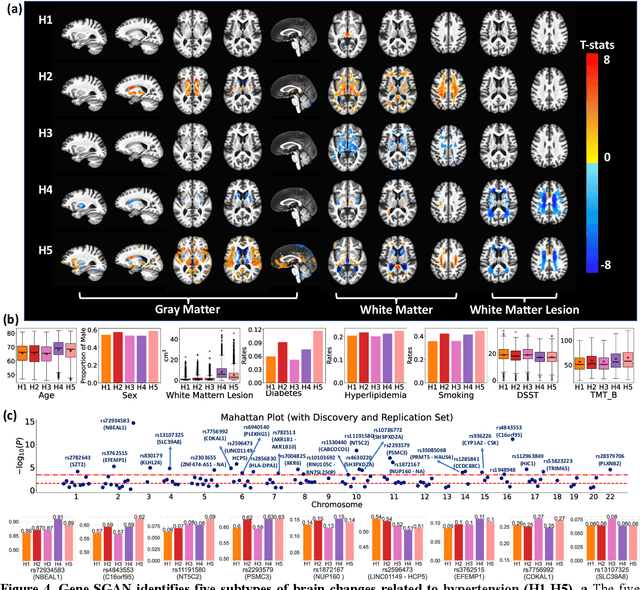
Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021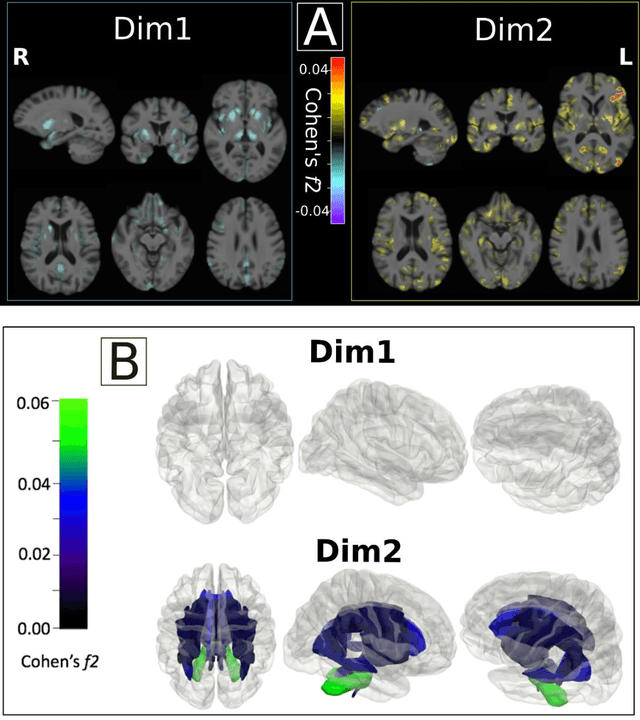
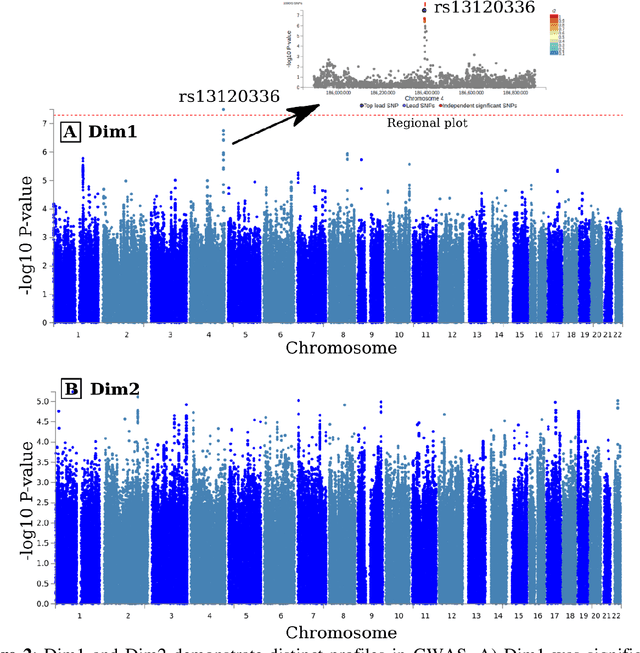
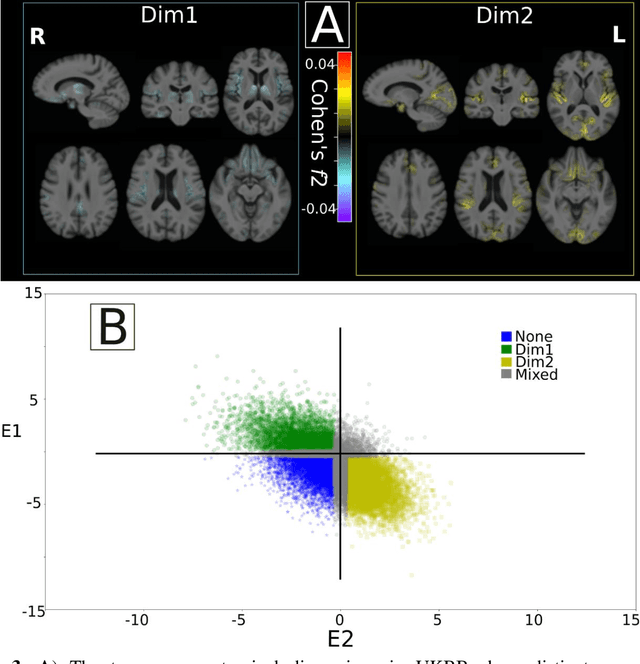
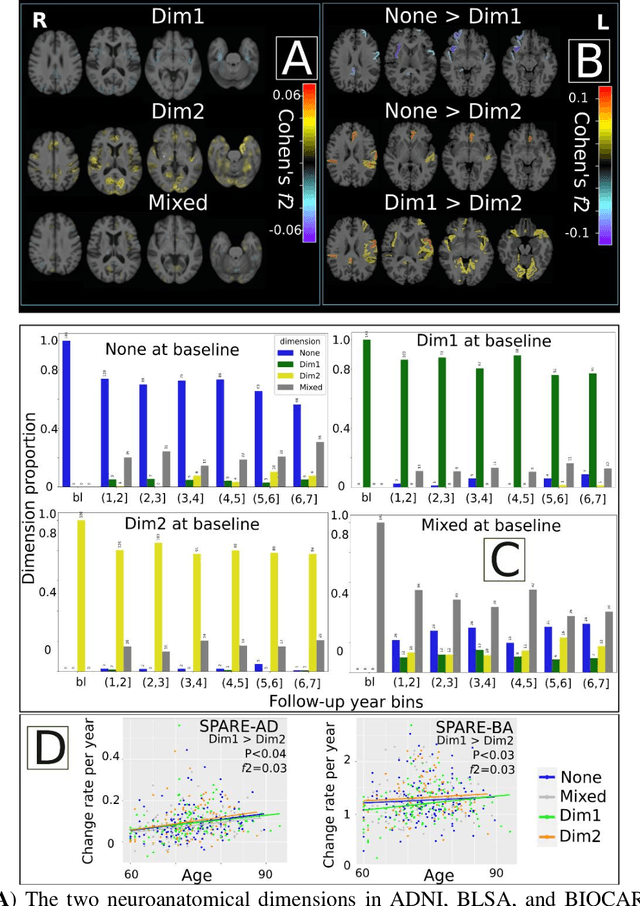
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge