Mohamad Habes
from the iSTAGING consortium, for the ADNI
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023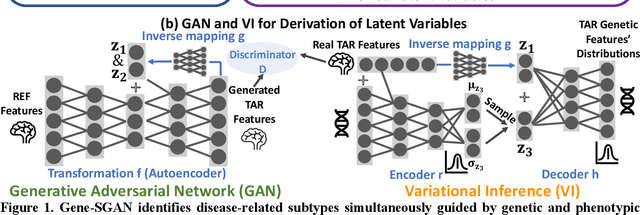
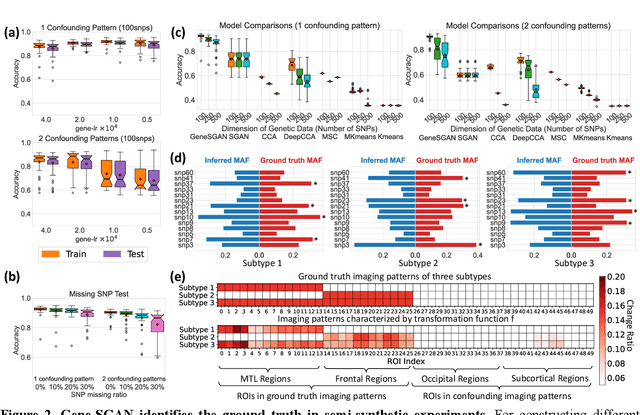
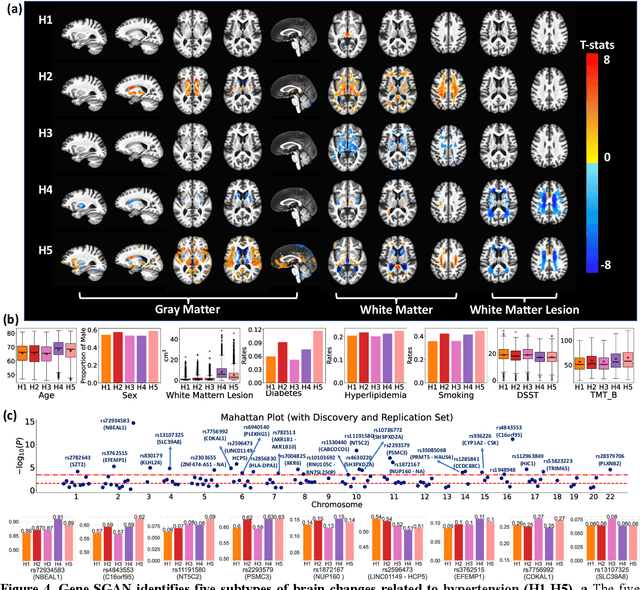
Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Deep Learning Based Detection of Enlarged Perivascular Spaces on Brain MRI
Sep 27, 2022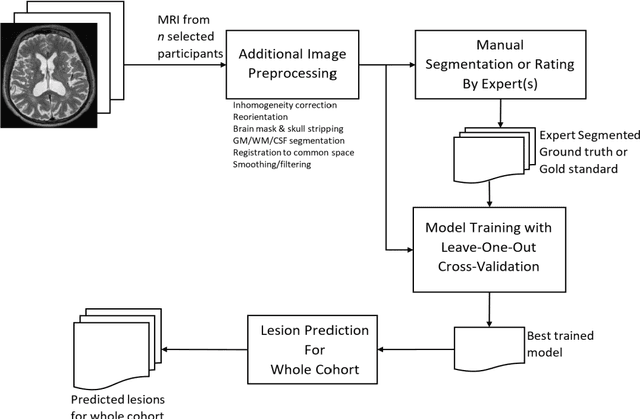
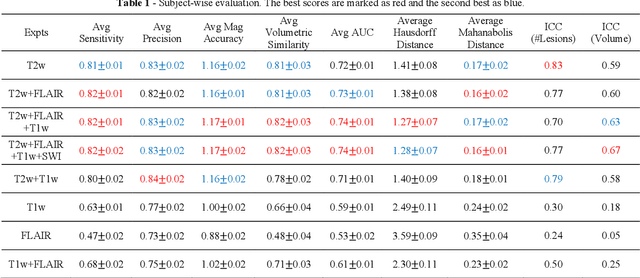
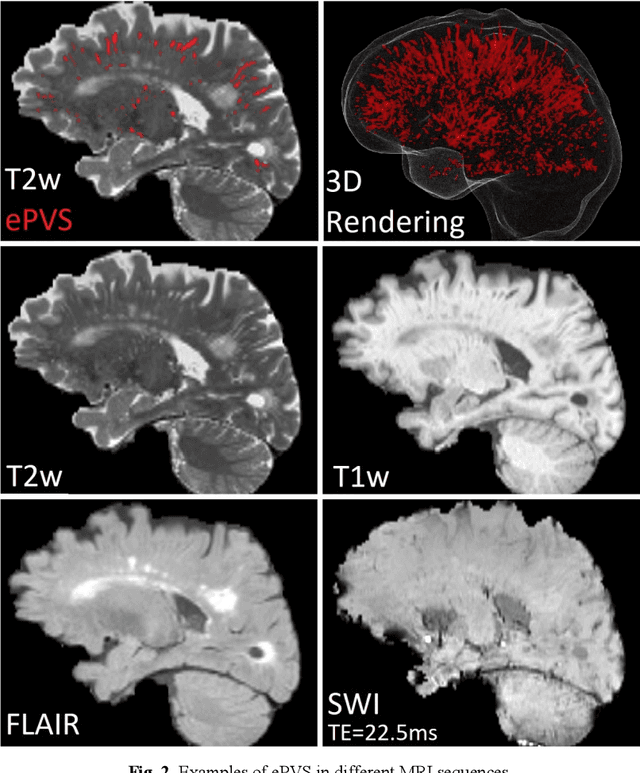
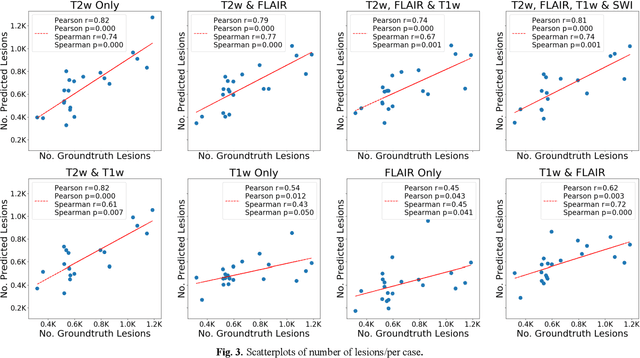
Abstract:Deep learning has been demonstrated effective in many neuroimaging applications. However, in many scenarios the number of imaging sequences capturing information related to small vessel disease lesions is insufficient to support data-driven techniques. Additionally, cohort-based studies may not always have the optimal or essential imaging sequences for accurate lesion detection. Therefore, it is necessary to determine which of these imaging sequences are essential for accurate detection. In this study we aimed to find the optimal combination of magnetic resonance imaging (MRI) sequences for deep learning-based detection of enlarged perivascular spaces (ePVS). To this end, we implemented an effective light-weight U-Net adapted for ePVS detection and comprehensively investigated different combinations of information from susceptibility weighted imaging (SWI), fluid-attenuated inversion recovery (FLAIR), T1-weighted (T1w) and T2-weighted (T2w) MRI sequences. We conclude that T2w MRI is the most important for accurate ePVS detection, and the incorporation of SWI, FLAIR and T1w MRI in the deep neural network could make insignificant improvements in accuracy.
Deep neural network heatmaps capture Alzheimer's disease patterns reported in a large meta-analysis of neuroimaging studies
Jul 22, 2022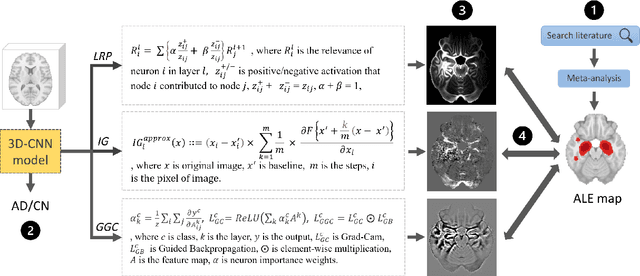
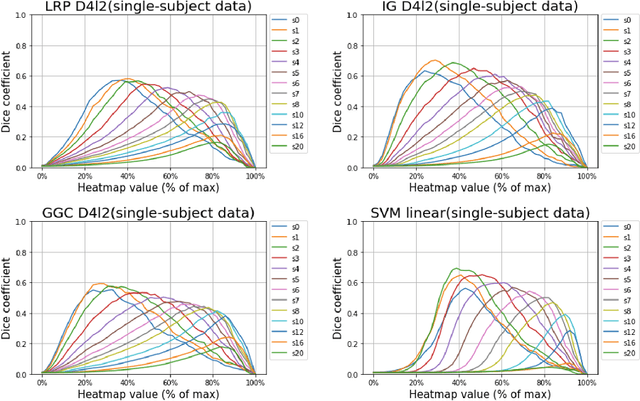
Abstract:Deep neural networks currently provide the most advanced and accurate machine learning models to distinguish between structural MRI scans of subjects with Alzheimer's disease and healthy controls. Unfortunately, the subtle brain alterations captured by these models are difficult to interpret because of the complexity of these multi-layer and non-linear models. Several heatmap methods have been proposed to address this issue and analyze the imaging patterns extracted from the deep neural networks, but no quantitative comparison between these methods has been carried out so far. In this work, we explore these questions by deriving heatmaps from Convolutional Neural Networks (CNN) trained using T1 MRI scans of the ADNI data set, and by comparing these heatmaps with brain maps corresponding to Support Vector Machines (SVM) coefficients. Three prominent heatmap methods are studied: Layer-wise Relevance Propagation (LRP), Integrated Gradients (IG), and Guided Grad-CAM (GGC). Contrary to prior studies where the quality of heatmaps was visually or qualitatively assessed, we obtained precise quantitative measures by computing overlap with a ground-truth map from a large meta-analysis that combined 77 voxel-based morphometry (VBM) studies independently from ADNI. Our results indicate that all three heatmap methods were able to capture brain regions covering the meta-analysis map and achieved better results than SVM coefficients. Among them, IG produced the heatmaps with the best overlap with the independent meta-analysis.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021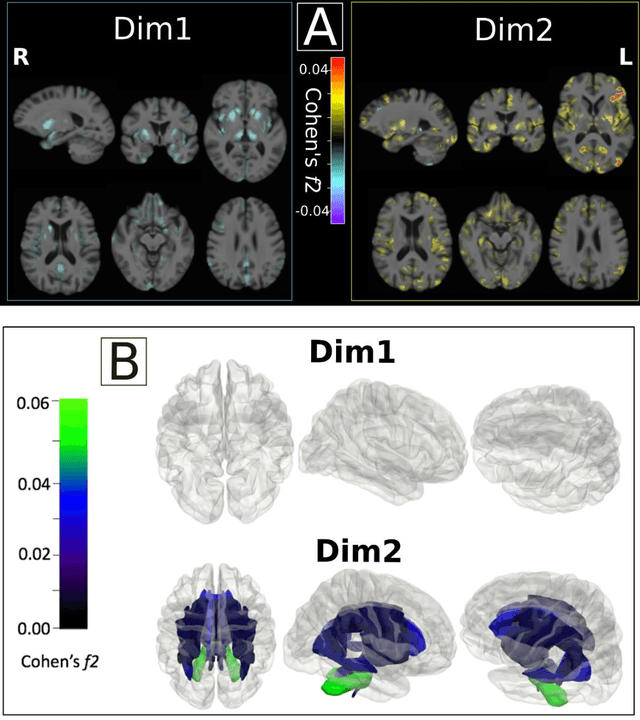
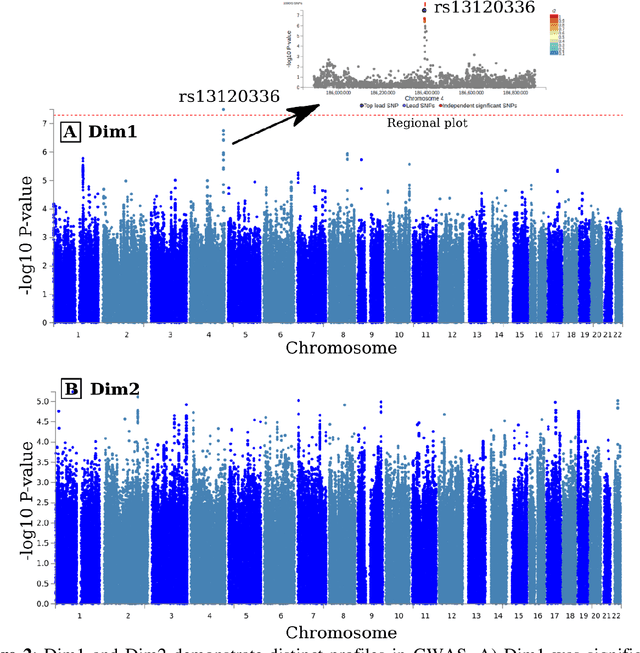
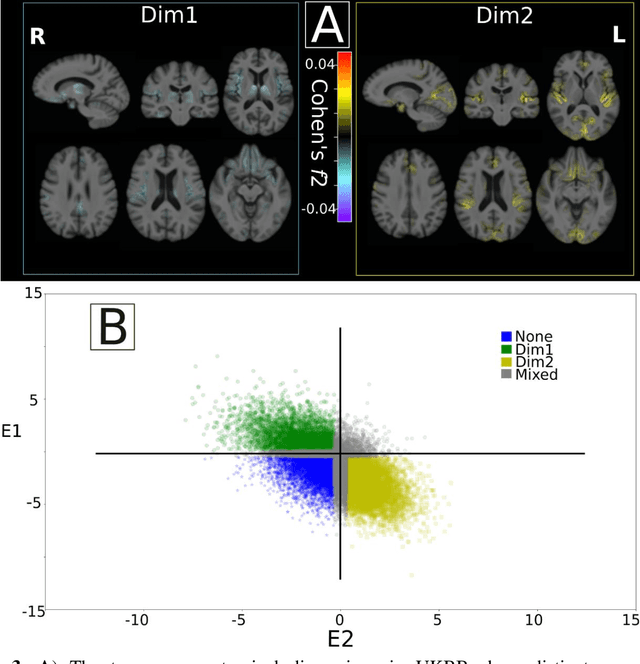
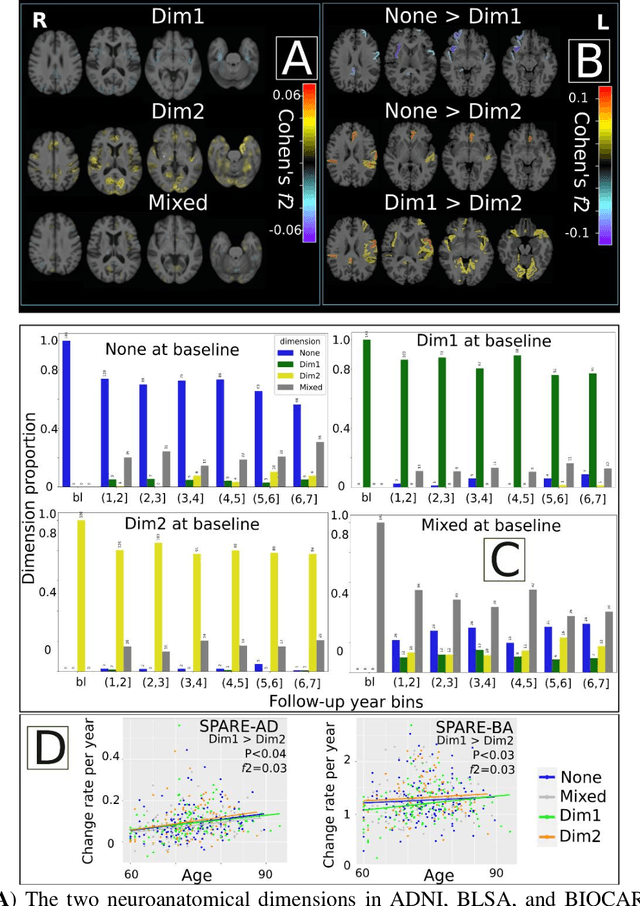
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
Disentangling Alzheimer's disease neurodegeneration from typical brain aging using machine learning
Sep 08, 2021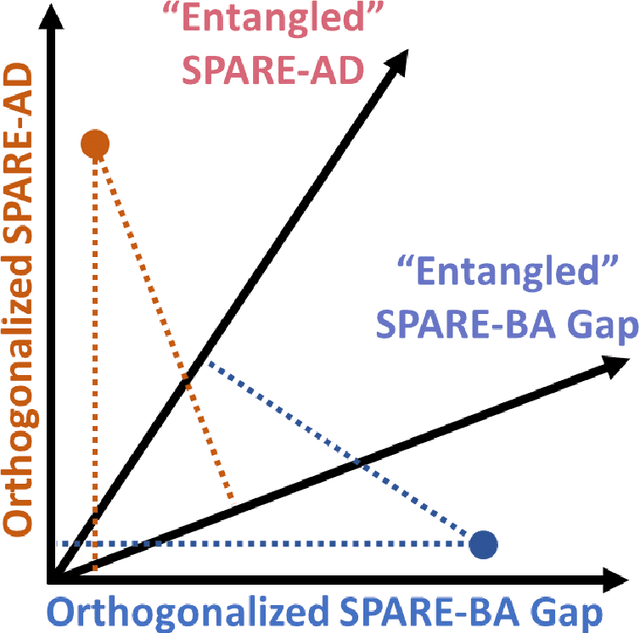
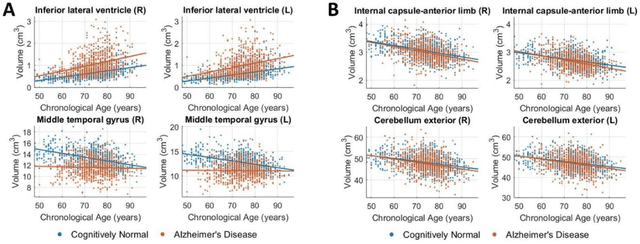
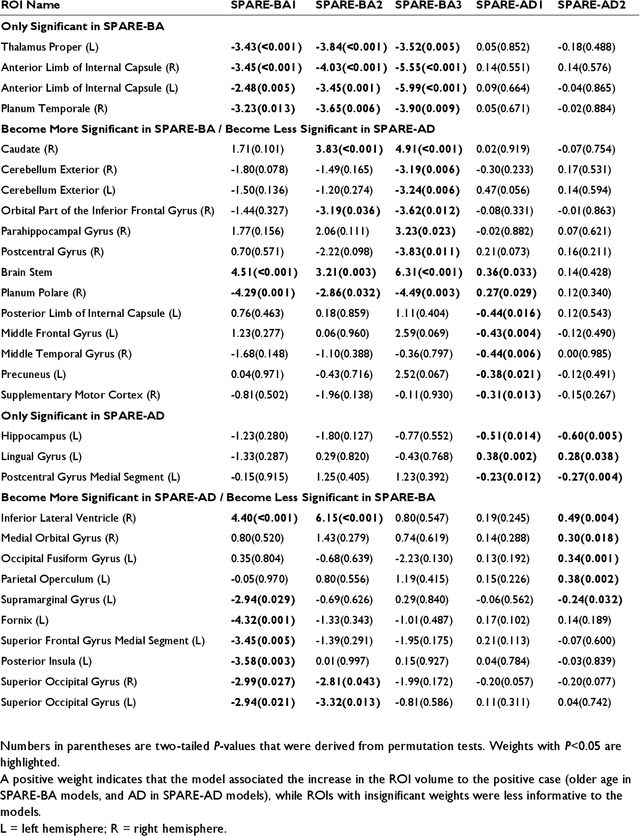
Abstract:Neuroimaging biomarkers that distinguish between typical brain aging and Alzheimer's disease (AD) are valuable for determining how much each contributes to cognitive decline. Machine learning models can derive multi-variate brain change patterns related to the two processes, including the SPARE-AD (Spatial Patterns of Atrophy for Recognition of Alzheimer's Disease) and SPARE-BA (of Brain Aging) investigated herein. However, substantial overlap between brain regions affected in the two processes confounds measuring them independently. We present a methodology toward disentangling the two. T1-weighted MRI images of 4,054 participants (48-95 years) with AD, mild cognitive impairment (MCI), or cognitively normal (CN) diagnoses from the iSTAGING (Imaging-based coordinate SysTem for AGIng and NeurodeGenerative diseases) consortium were analyzed. First, a subset of AD patients and CN adults were selected based purely on clinical diagnoses to train SPARE-BA1 (regression of age using CN individuals) and SPARE-AD1 (classification of CN versus AD). Second, analogous groups were selected based on clinical and molecular markers to train SPARE-BA2 and SPARE-AD2: amyloid-positive (A+) AD continuum group (consisting of A+AD, A+MCI, and A+ and tau-positive CN individuals) and amyloid-negative (A-) CN group. Finally, the combined group of the AD continuum and A-/CN individuals was used to train SPARE-BA3, with the intention to estimate brain age regardless of AD-related brain changes. Disentangled SPARE models derived brain patterns that were more specific to the two types of the brain changes. Correlation between the SPARE-BA and SPARE-AD was significantly reduced. Correlation of disentangled SPARE-AD was non-inferior to the molecular measurements and to the number of APOE4 alleles, but was less to AD-related psychometric test scores, suggesting contribution of advanced brain aging to these scores.
Disentangling brain heterogeneity via semi-supervised deep-learning and MRI: dimensional representations of Alzheimer's Disease
Feb 24, 2021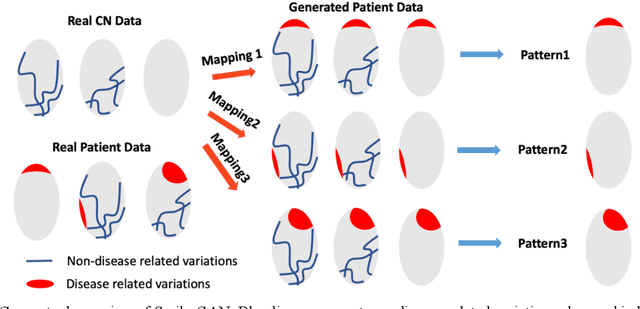
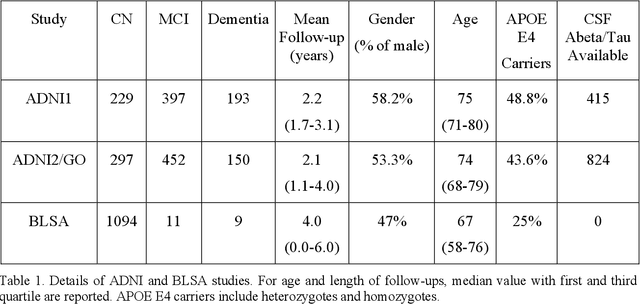
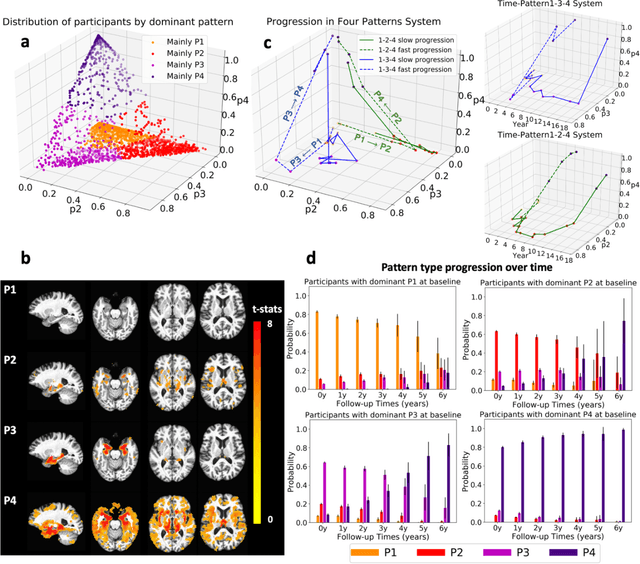
Abstract:Heterogeneity of brain diseases is a challenge for precision diagnosis/prognosis. We describe and validate Smile-GAN (SeMI-supervised cLustEring-Generative Adversarial Network), a novel semi-supervised deep-clustering method, which dissects neuroanatomical heterogeneity, enabling identification of disease subtypes via their imaging signatures relative to controls. When applied to MRIs (2 studies; 2,832 participants; 8,146 scans) including cognitively normal individuals and those with cognitive impairment and dementia, Smile-GAN identified 4 neurodegenerative patterns/axes: P1, normal anatomy and highest cognitive performance; P2, mild/diffuse atrophy and more prominent executive dysfunction; P3, focal medial temporal atrophy and relatively greater memory impairment; P4, advanced neurodegeneration. Further application to longitudinal data revealed two distinct progression pathways: P1$\rightarrow$P2$\rightarrow$P4 and P1$\rightarrow$P3$\rightarrow$P4. Baseline expression of these patterns predicted the pathway and rate of future neurodegeneration. Pattern expression offered better yet complementary performance in predicting clinical progression, compared to amyloid/tau. These deep-learning derived biomarkers offer promise for precision diagnostics and targeted clinical trial recruitment.
Medical Image Harmonization Using Deep Learning Based Canonical Mapping: Toward Robust and Generalizable Learning in Imaging
Oct 11, 2020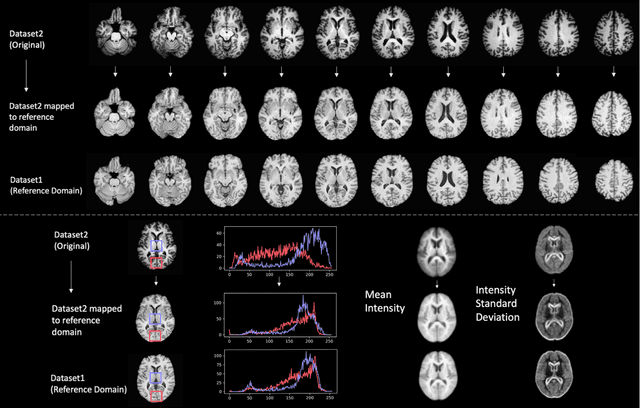
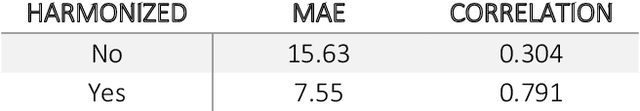
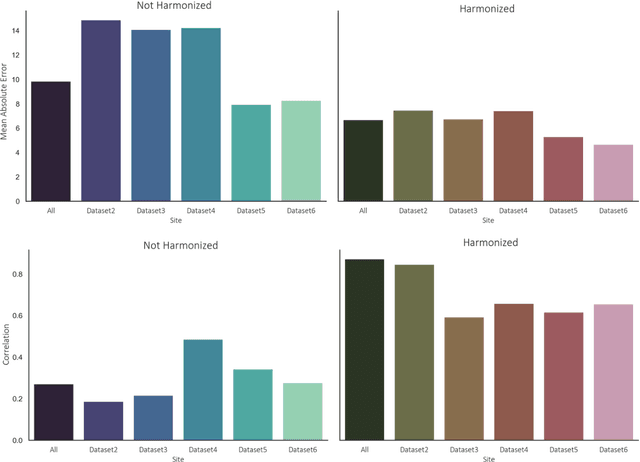
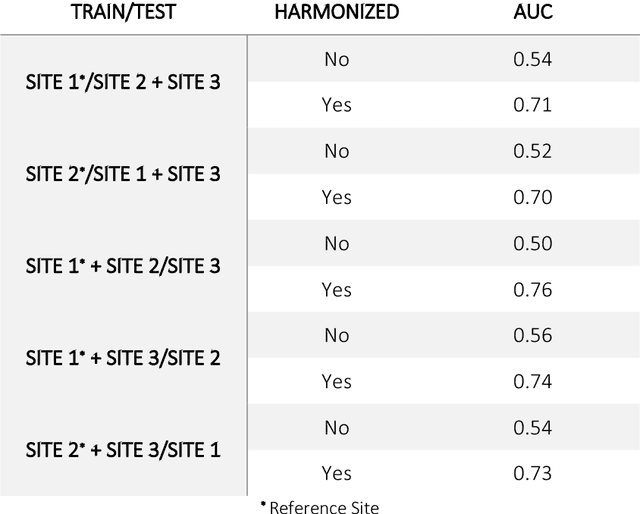
Abstract:Conventional and deep learning-based methods have shown great potential in the medical imaging domain, as means for deriving diagnostic, prognostic, and predictive biomarkers, and by contributing to precision medicine. However, these methods have yet to see widespread clinical adoption, in part due to limited generalization performance across various imaging devices, acquisition protocols, and patient populations. In this work, we propose a new paradigm in which data from a diverse range of acquisition conditions are "harmonized" to a common reference domain, where accurate model learning and prediction can take place. By learning an unsupervised image to image canonical mapping from diverse datasets to a reference domain using generative deep learning models, we aim to reduce confounding data variation while preserving semantic information, thereby rendering the learning task easier in the reference domain. We test this approach on two example problems, namely MRI-based brain age prediction and classification of schizophrenia, leveraging pooled cohorts of neuroimaging MRI data spanning 9 sites and 9701 subjects. Our results indicate a substantial improvement in these tasks in out-of-sample data, even when training is restricted to a single site.
DEEPMIR: A DEEP convolutional neural network for differential detection of cerebral Microbleeds and IRon deposits in MRI
Sep 30, 2020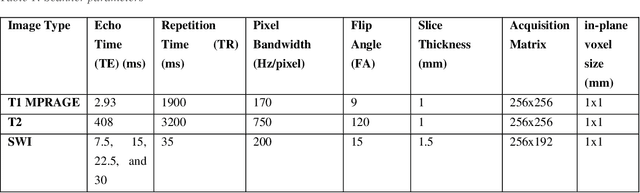
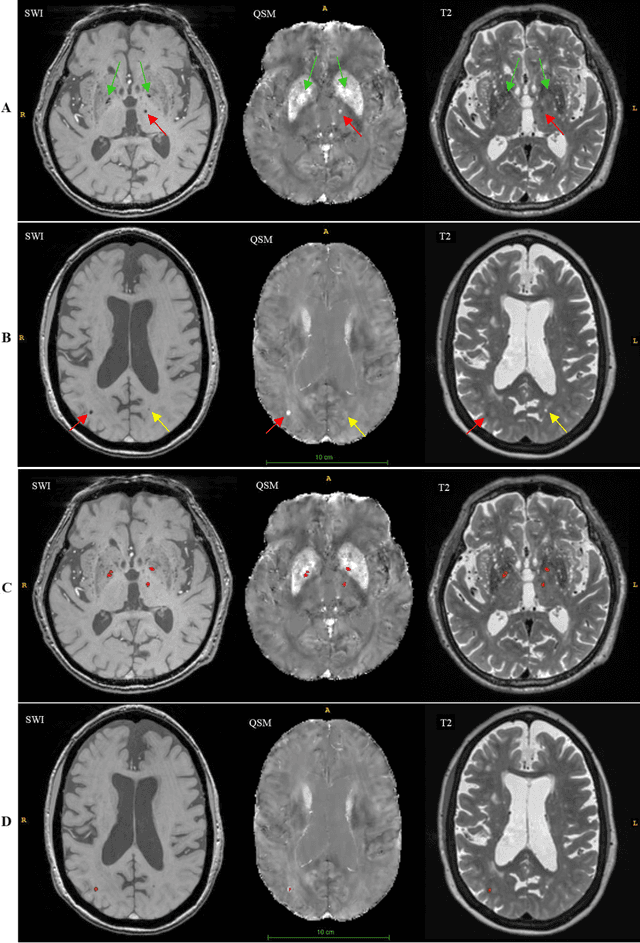
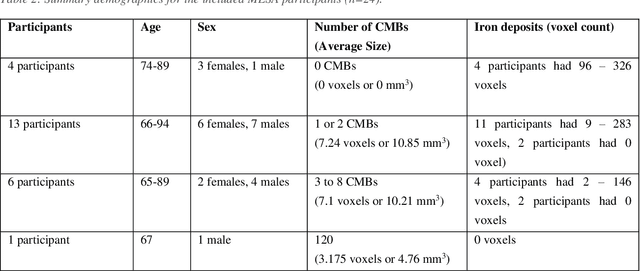

Abstract:Background: Cerebral microbleeds (CMBs) and non-hemorrhage iron deposits in the basal ganglia have been associated with brain aging, vascular disease and neurodegenerative disorders. Recent advances using quantitative susceptibility mapping (QSM) make it possible to differentiate iron content from mineralization in-vivo using magnetic resonance imaging (MRI). However, automated detection of such lesions is still challenging, making quantification in large cohort bases studies rather limited. Purpose: Development of a fully automated method using deep learning for detecting CMBs and basal ganglia iron deposits using multimodal MRI. Materials and Methods: We included a convenience sample of 24 participants from the MESA cohort and used T2-weighted images, susceptibility weighted imaging (SWI), and QSM to segment the lesions. We developed a protocol for simultaneous manual annotation of CMBs and non-hemorrhage iron deposits in the basal ganglia, which resulted in defining the gold standard. This gold standard was then used to train a deep convolution neural network (CNN) model. Specifically, we adapted the U-Net model with a higher number of resolution layers to be able to detect small lesions such as CMBs from standard resolution MRI which are used in cohort-based studies. The detection performance was then evaluated using the cross-validation principle in order to ensure generalization of the results. Results: With multi-class CNN models, we achieved an average sensitivity and precision of about 0.8 and 0.6, respectively for detecting CMBs. The same framework detected non-hemorrhage iron deposits reaching an average sensitivity and precision of about 0.8. Conclusions: Our results showed that deep learning could automate the detection of small vessel disease lesions and including multimodal MR data such as QSM can improve the detection of CMB and non-hemorrhage iron deposits.
DeepMRSeg: A convolutional deep neural network for anatomy and abnormality segmentation on MR images
Jul 03, 2019

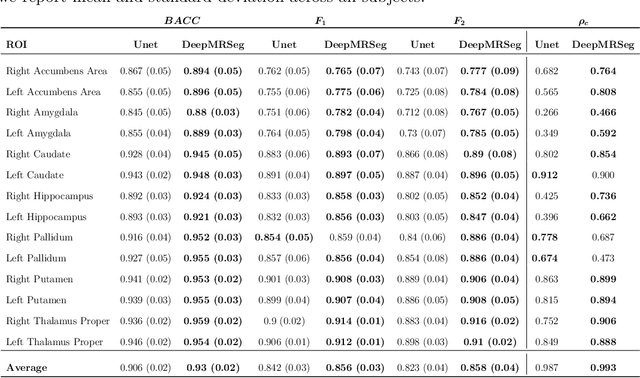

Abstract:Segmentation has been a major task in neuroimaging. A large number of automated methods have been developed for segmenting healthy and diseased brain tissues. In recent years, deep learning techniques have attracted a lot of attention as a result of their high accuracy in different segmentation problems. We present a new deep learning based segmentation method, DeepMRSeg, that can be applied in a generic way to a variety of segmentation tasks. The proposed architecture combines recent advances in the field of biomedical image segmentation and computer vision. We use a modified UNet architecture that takes advantage of multiple convolution filter sizes to achieve multi-scale feature extraction adaptive to the desired segmentation task. Importantly, our method operates on minimally processed raw MRI scan. We validated our method on a wide range of segmentation tasks, including white matter lesion segmentation, segmentation of deep brain structures and hippocampus segmentation. We provide code and pre-trained models to allow researchers apply our method on their own datasets.
A deep learning model for early prediction of Alzheimer's disease dementia based on hippocampal MRI
Apr 15, 2019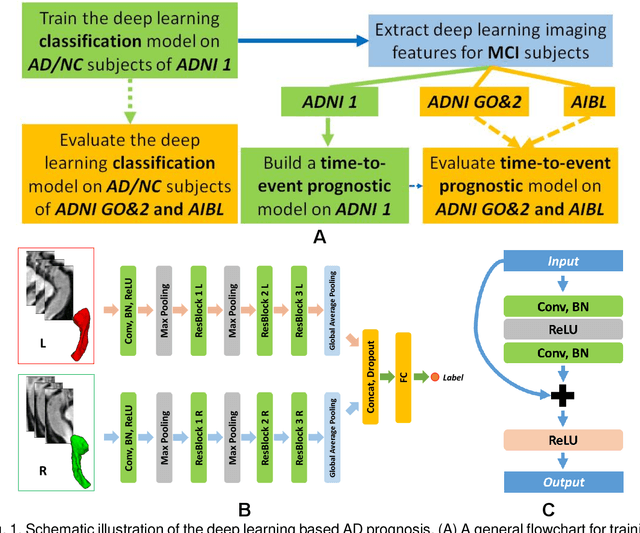


Abstract:Introduction: It is challenging at baseline to predict when and which individuals who meet criteria for mild cognitive impairment (MCI) will ultimately progress to Alzheimer's disease (AD) dementia. Methods: A deep learning method is developed and validated based on MRI scans of 2146 subjects (803 for training and 1343 for validation) to predict MCI subjects' progression to AD dementia in a time-to-event analysis setting. Results: The deep learning time-to-event model predicted individual subjects' progression to AD dementia with a concordance index (C-index) of 0.762 on 439 ADNI testing MCI subjects with follow-up duration from 6 to 78 months (quartiles: [24, 42, 54]) and a C-index of 0.781 on 40 AIBL testing MCI subjects with follow-up duration from 18-54 months (quartiles: [18, 36,54]). The predicted progression risk also clustered individual subjects into subgroups with significant differences in their progression time to AD dementia (p<0.0002). Improved performance for predicting progression to AD dementia (C-index=0.864) was obtained when the deep learning based progression risk was combined with baseline clinical measures. Conclusion: Our method provides a cost effective and accurate means for prognosis and potentially to facilitate enrollment in clinical trials with individuals likely to progress within a specific temporal period.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge