Disentangling brain heterogeneity via semi-supervised deep-learning and MRI: dimensional representations of Alzheimer's Disease
Paper and Code
Feb 24, 2021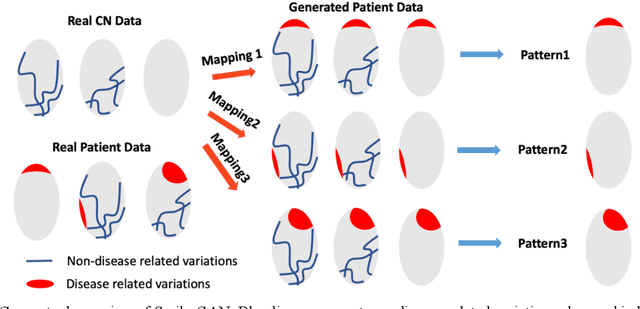
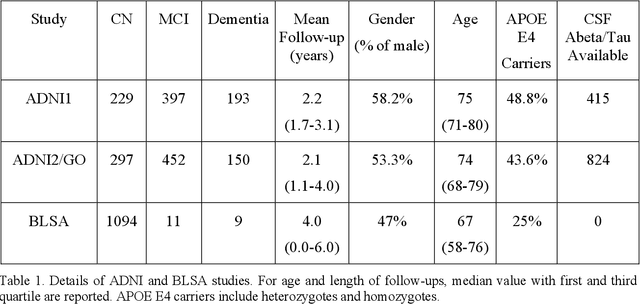
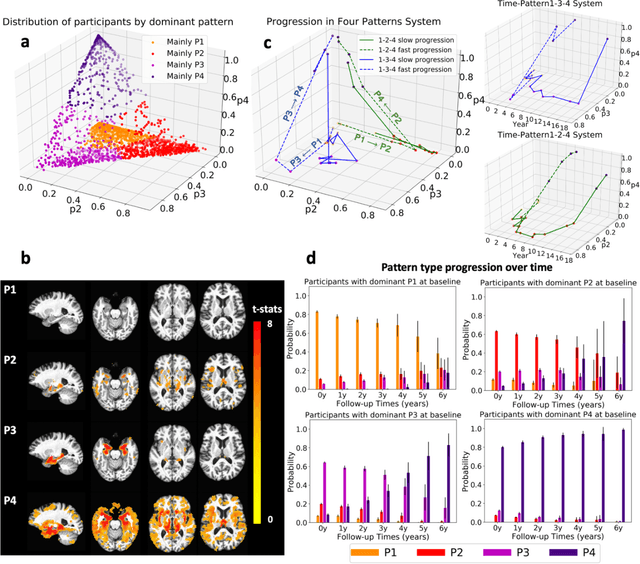
Heterogeneity of brain diseases is a challenge for precision diagnosis/prognosis. We describe and validate Smile-GAN (SeMI-supervised cLustEring-Generative Adversarial Network), a novel semi-supervised deep-clustering method, which dissects neuroanatomical heterogeneity, enabling identification of disease subtypes via their imaging signatures relative to controls. When applied to MRIs (2 studies; 2,832 participants; 8,146 scans) including cognitively normal individuals and those with cognitive impairment and dementia, Smile-GAN identified 4 neurodegenerative patterns/axes: P1, normal anatomy and highest cognitive performance; P2, mild/diffuse atrophy and more prominent executive dysfunction; P3, focal medial temporal atrophy and relatively greater memory impairment; P4, advanced neurodegeneration. Further application to longitudinal data revealed two distinct progression pathways: P1$\rightarrow$P2$\rightarrow$P4 and P1$\rightarrow$P3$\rightarrow$P4. Baseline expression of these patterns predicted the pathway and rate of future neurodegeneration. Pattern expression offered better yet complementary performance in predicting clinical progression, compared to amyloid/tau. These deep-learning derived biomarkers offer promise for precision diagnostics and targeted clinical trial recruitment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge