Nicolas Honnorat
Deep neural network heatmaps capture Alzheimer's disease patterns reported in a large meta-analysis of neuroimaging studies
Jul 22, 2022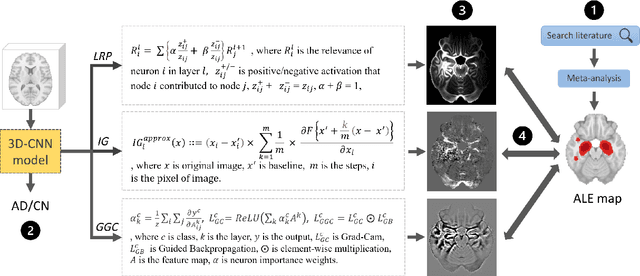
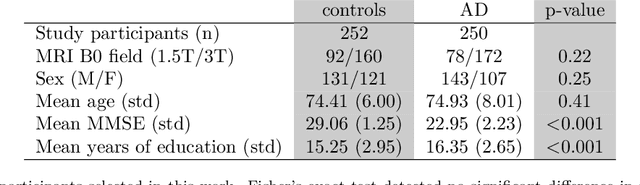
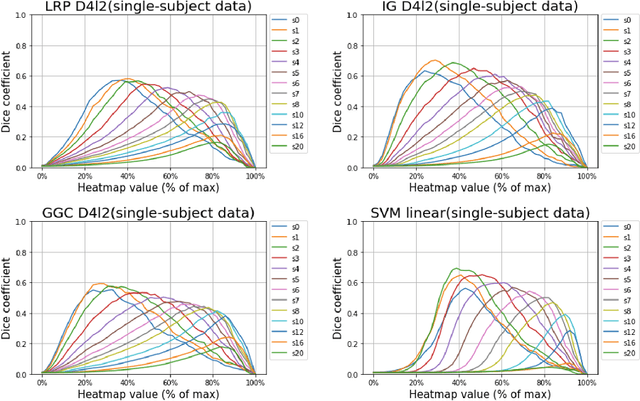
Abstract:Deep neural networks currently provide the most advanced and accurate machine learning models to distinguish between structural MRI scans of subjects with Alzheimer's disease and healthy controls. Unfortunately, the subtle brain alterations captured by these models are difficult to interpret because of the complexity of these multi-layer and non-linear models. Several heatmap methods have been proposed to address this issue and analyze the imaging patterns extracted from the deep neural networks, but no quantitative comparison between these methods has been carried out so far. In this work, we explore these questions by deriving heatmaps from Convolutional Neural Networks (CNN) trained using T1 MRI scans of the ADNI data set, and by comparing these heatmaps with brain maps corresponding to Support Vector Machines (SVM) coefficients. Three prominent heatmap methods are studied: Layer-wise Relevance Propagation (LRP), Integrated Gradients (IG), and Guided Grad-CAM (GGC). Contrary to prior studies where the quality of heatmaps was visually or qualitatively assessed, we obtained precise quantitative measures by computing overlap with a ground-truth map from a large meta-analysis that combined 77 voxel-based morphometry (VBM) studies independently from ADNI. Our results indicate that all three heatmap methods were able to capture brain regions covering the meta-analysis map and achieved better results than SVM coefficients. Among them, IG produced the heatmaps with the best overlap with the independent meta-analysis.
Variational AutoEncoder For Regression: Application to Brain Aging Analysis
Apr 11, 2019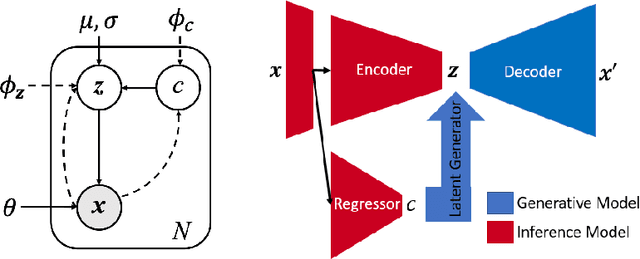
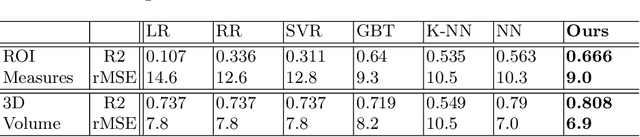
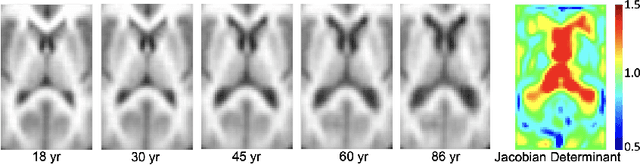
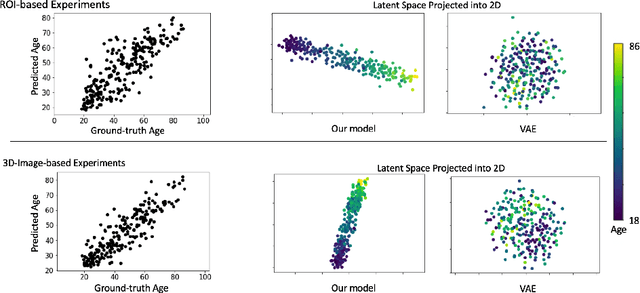
Abstract:While unsupervised variational autoencoders (VAE) have become a powerful tool in neuroimage analysis, their application to supervised learning is under-explored. We aim to close this gap by proposing a unified probabilistic model for learning the latent space of imaging data and performing supervised regression. Based on recent advances in learning disentangled representations, the novel generative process explicitly models the conditional distribution of latent representations with respect to the regression target variable. Performing a variational inference procedure on this model leads to joint regularization between the VAE and a neural-network regressor. In predicting the age of 245 subjects from their structural Magnetic Resonance (MR) images, our model is more accurate than state-of-the-art methods when applied to either region-of-interest (ROI) measurements or raw 3D volume images. More importantly, unlike simple feed-forward neural-networks, disentanglement of age in latent representations allows for intuitive interpretation of the structural developmental patterns of the human brain.
Truncated Gaussian-Mixture Variational AutoEncoder
Mar 08, 2019



Abstract:Variation Autoencoder (VAE) has become a powerful tool in modeling the non-linear generative process of data from a low-dimensional latent space. Recently, several studies have proposed to use VAE for unsupervised clustering by using mixture models to capture the multi-modal structure of latent representations. This strategy, however, is ineffective when there are outlier data samples whose latent representations are meaningless, yet contaminating the estimation of key major clusters in the latent space. This exact problem arises in the context of resting-state fMRI (rs-fMRI) analysis, where clustering major functional connectivity patterns is often hindered by heavy noise of rs-fMRI and many minor clusters (rare connectivity patterns) of no interest to analysis. In this paper we propose a novel generative process, in which we use a Gaussian-mixture to model a few major clusters in the data, and use a non-informative uniform distribution to capture the remaining data. We embed this truncated Gaussian-Mixture model in a Variational AutoEncoder framework to obtain a general joint clustering and outlier detection approach, called tGM-VAE. We demonstrated the applicability of tGM-VAE on the MNIST dataset and further validated it in the context of rs-fMRI connectivity analysis.
Sparse Hierachical Extrapolated Parametric Methods for Cortical Data Analysis
Apr 27, 2017



Abstract:Many neuroimaging studies focus on the cortex, in order to benefit from better signal to noise ratios and reduced computational burden. Cortical data are usually projected onto a reference mesh, where subsequent analyses are carried out. Several multiscale approaches have been proposed for analyzing these surface data, such as spherical harmonics and graph wavelets. As far as we know, however, the hierarchical structure of the template icosahedral meshes used by most neuroimaging software has never been exploited for cortical data factorization. In this paper, we demonstrate how the structure of the ubiquitous icosahedral meshes can be exploited by data factorization methods such as sparse dictionary learning, and we assess the optimization speed-up offered by extrapolation methods in this context. By testing different sparsity-inducing norms, extrapolation methods, and factorization schemes, we compare the performances of eleven methods for analyzing four datasets: two structural and two functional MRI datasets obtained by processing the data publicly available for the hundred unrelated subjects of the Human Connectome Project. Our results demonstrate that, depending on the level of details requested, a speedup of several orders of magnitudes can be obtained.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge