Kerstin Ritter
Brain-Semantoks: Learning Semantic Tokens of Brain Dynamics with a Self-Distilled Foundation Model
Dec 12, 2025Abstract:The development of foundation models for functional magnetic resonance imaging (fMRI) time series holds significant promise for predicting phenotypes related to disease and cognition. Current models, however, are often trained using a mask-and-reconstruct objective on small brain regions. This focus on low-level information leads to representations that are sensitive to noise and temporal fluctuations, necessitating extensive fine-tuning for downstream tasks. We introduce Brain-Semantoks, a self-supervised framework designed specifically to learn abstract representations of brain dynamics. Its architecture is built on two core innovations: a semantic tokenizer that aggregates noisy regional signals into robust tokens representing functional networks, and a self-distillation objective that enforces representational stability across time. We show that this objective is stabilized through a novel training curriculum, ensuring the model robustly learns meaningful features from low signal-to-noise time series. We demonstrate that learned representations enable strong performance on a variety of downstream tasks even when only using a linear probe. Furthermore, we provide comprehensive scaling analyses indicating more unlabeled data reliably results in out-of-distribution performance gains without domain adaptation.
Identifying confounders in deep-learning-based model predictions using DeepRepViz
Sep 27, 2023Abstract:Deep Learning (DL) models are increasingly used to analyze neuroimaging data and uncover insights about the brain, brain pathologies, and psychological traits. However, extraneous `confounders' variables such as the age of the participants, sex, or imaging artifacts can bias model predictions, preventing the models from learning relevant brain-phenotype relationships. In this study, we provide a solution called the `DeepRepViz' framework that enables researchers to systematically detect confounders in their DL model predictions. The framework consists of (1) a metric that quantifies the effect of potential confounders and (2) a visualization tool that allows researchers to qualitatively inspect what the DL model is learning. By performing experiments on simulated and neuroimaging datasets, we demonstrate the benefits of using DeepRepViz in combination with DL models. For example, experiments on the neuroimaging datasets reveal that sex is a significant confounder in a DL model predicting chronic alcohol users (Con-score=0.35). Similarly, DeepRepViz identifies age as a confounder in a DL model predicting participants' performance on a cognitive task (Con-score=0.3). Overall, DeepRepViz enables researchers to systematically test for potential confounders and expose DL models that rely on extraneous information such as age, sex, or imaging artifacts.
Benchmark data to study the influence of pre-training on explanation performance in MR image classification
Jun 21, 2023



Abstract:Convolutional Neural Networks (CNNs) are frequently and successfully used in medical prediction tasks. They are often used in combination with transfer learning, leading to improved performance when training data for the task are scarce. The resulting models are highly complex and typically do not provide any insight into their predictive mechanisms, motivating the field of 'explainable' artificial intelligence (XAI). However, previous studies have rarely quantitatively evaluated the 'explanation performance' of XAI methods against ground-truth data, and transfer learning and its influence on objective measures of explanation performance has not been investigated. Here, we propose a benchmark dataset that allows for quantifying explanation performance in a realistic magnetic resonance imaging (MRI) classification task. We employ this benchmark to understand the influence of transfer learning on the quality of explanations. Experimental results show that popular XAI methods applied to the same underlying model differ vastly in performance, even when considering only correctly classified examples. We further observe that explanation performance strongly depends on the task used for pre-training and the number of CNN layers pre-trained. These results hold after correcting for a substantial correlation between explanation and classification performance.
Promises and pitfalls of deep neural networks in neuroimaging-based psychiatric research
Jan 20, 2023



Abstract:By promising more accurate diagnostics and individual treatment recommendations, deep neural networks and in particular convolutional neural networks have advanced to a powerful tool in medical imaging. Here, we first give an introduction into methodological key concepts and resulting methodological promises including representation and transfer learning, as well as modelling domain-specific priors. After reviewing recent applications within neuroimaging-based psychiatric research, such as the diagnosis of psychiatric diseases, delineation of disease subtypes, normative modeling, and the development of neuroimaging biomarkers, we discuss current challenges. This includes for example the difficulty of training models on small, heterogeneous and biased data sets, the lack of validity of clinical labels, algorithmic bias, and the influence of confounding variables.
Deep neural network heatmaps capture Alzheimer's disease patterns reported in a large meta-analysis of neuroimaging studies
Jul 22, 2022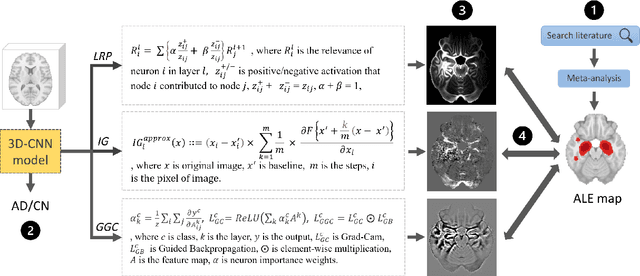
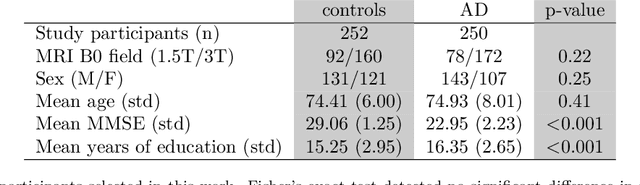
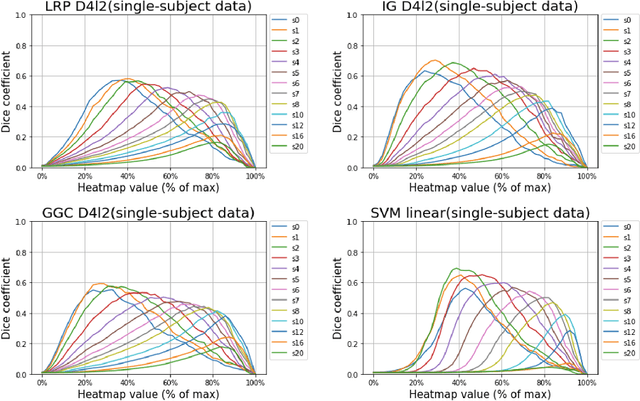
Abstract:Deep neural networks currently provide the most advanced and accurate machine learning models to distinguish between structural MRI scans of subjects with Alzheimer's disease and healthy controls. Unfortunately, the subtle brain alterations captured by these models are difficult to interpret because of the complexity of these multi-layer and non-linear models. Several heatmap methods have been proposed to address this issue and analyze the imaging patterns extracted from the deep neural networks, but no quantitative comparison between these methods has been carried out so far. In this work, we explore these questions by deriving heatmaps from Convolutional Neural Networks (CNN) trained using T1 MRI scans of the ADNI data set, and by comparing these heatmaps with brain maps corresponding to Support Vector Machines (SVM) coefficients. Three prominent heatmap methods are studied: Layer-wise Relevance Propagation (LRP), Integrated Gradients (IG), and Guided Grad-CAM (GGC). Contrary to prior studies where the quality of heatmaps was visually or qualitatively assessed, we obtained precise quantitative measures by computing overlap with a ground-truth map from a large meta-analysis that combined 77 voxel-based morphometry (VBM) studies independently from ADNI. Our results indicate that all three heatmap methods were able to capture brain regions covering the meta-analysis map and achieved better results than SVM coefficients. Among them, IG produced the heatmaps with the best overlap with the independent meta-analysis.
Feature visualization for convolutional neural network models trained on neuroimaging data
Mar 24, 2022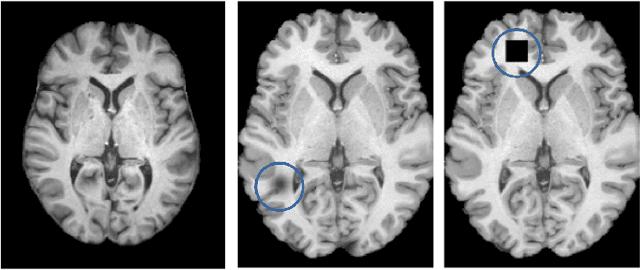
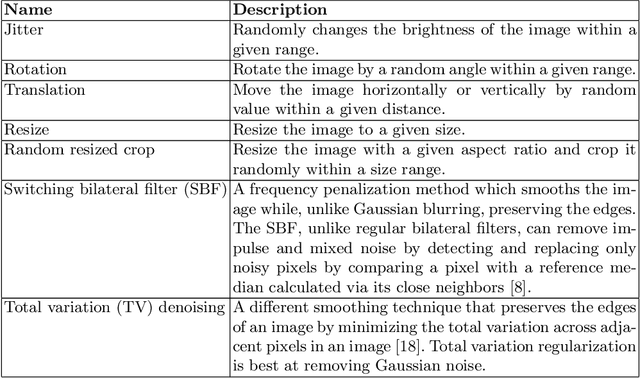
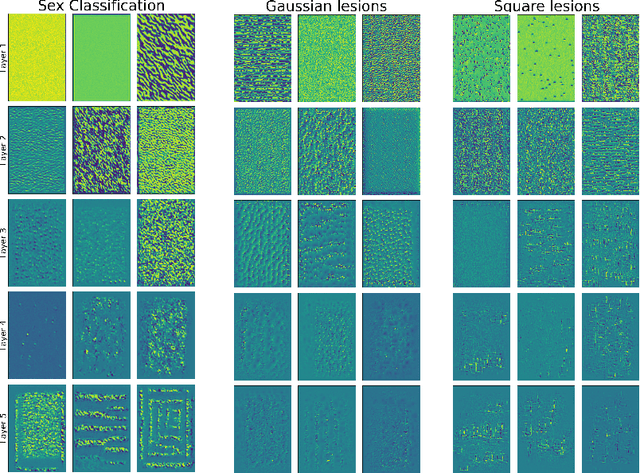
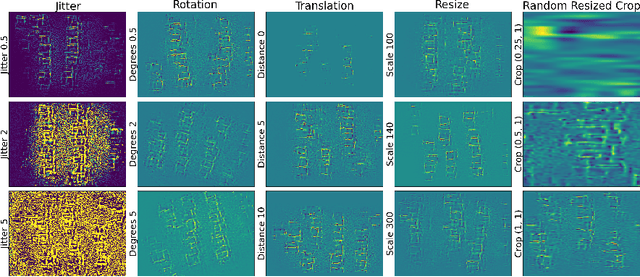
Abstract:A major prerequisite for the application of machine learning models in clinical decision making is trust and interpretability. Current explainability studies in the neuroimaging community have mostly focused on explaining individual decisions of trained models, e.g. obtained by a convolutional neural network (CNN). Using attribution methods such as layer-wise relevance propagation or SHAP heatmaps can be created that highlight which regions of an input are more relevant for the decision than others. While this allows the detection of potential data set biases and can be used as a guide for a human expert, it does not allow an understanding of the underlying principles the model has learned. In this study, we instead show, to the best of our knowledge, for the first time results using feature visualization of neuroimaging CNNs. Particularly, we have trained CNNs for different tasks including sex classification and artificial lesion classification based on structural magnetic resonance imaging (MRI) data. We have then iteratively generated images that maximally activate specific neurons, in order to visualize the patterns they respond to. To improve the visualizations we compared several regularization strategies. The resulting images reveal the learned concepts of the artificial lesions, including their shapes, but remain hard to interpret for abstract features in the sex classification task.
Evaluating saliency methods on artificial data with different background types
Dec 09, 2021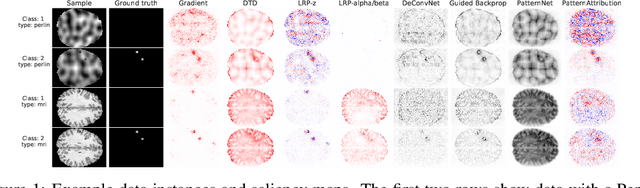
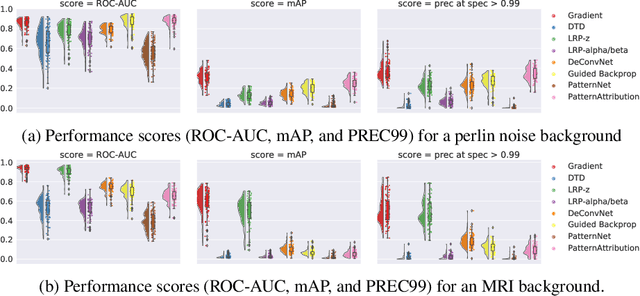
Abstract:Over the last years, many 'explainable artificial intelligence' (xAI) approaches have been developed, but these have not always been objectively evaluated. To evaluate the quality of heatmaps generated by various saliency methods, we developed a framework to generate artificial data with synthetic lesions and a known ground truth map. Using this framework, we evaluated two data sets with different backgrounds, Perlin noise and 2D brain MRI slices, and found that the heatmaps vary strongly between saliency methods and backgrounds. We strongly encourage further evaluation of saliency maps and xAI methods using this framework before applying these in clinical or other safety-critical settings.
Harnessing spatial homogeneity of neuroimaging data: patch individual filter layers for CNNs
Jul 23, 2020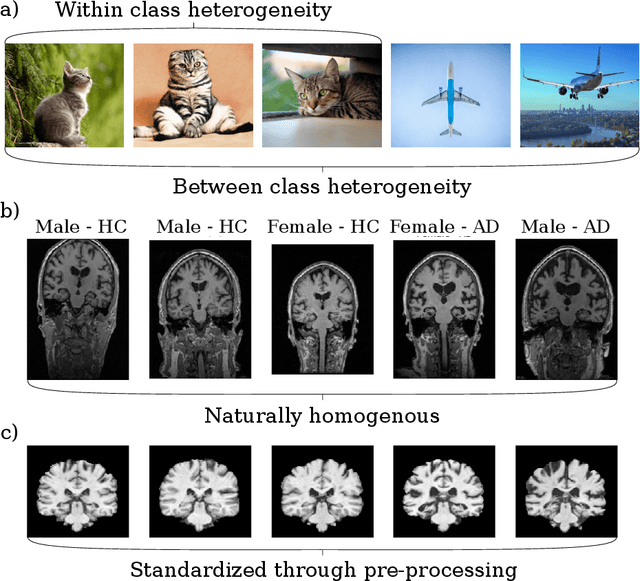
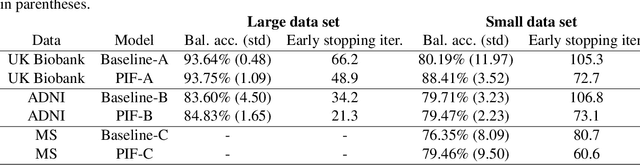
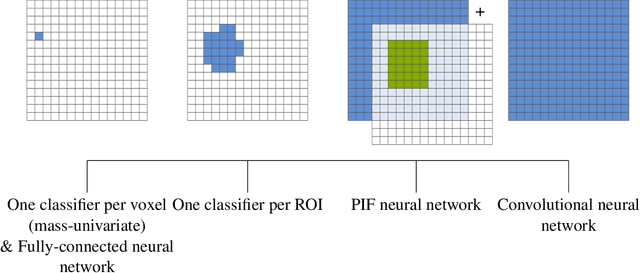
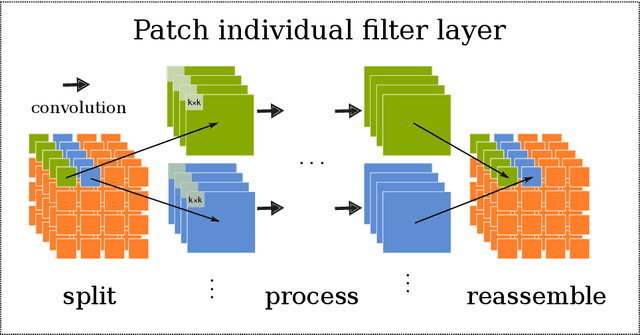
Abstract:Neuroimaging data, e.g. obtained from magnetic resonance imaging (MRI), is comparably homogeneous due to (1) the uniform structure of the brain and (2) additional efforts to spatially normalize the data to a standard template using linear and non-linear transformations. Convolutional neural networks (CNNs), in contrast, have been specifically designed for highly heterogeneous data, such as natural images, by sliding convolutional filters over different positions in an image. Here, we suggest a new CNN architecture that combines the idea of hierarchical abstraction in neural networks with a prior on the spatial homogeneity of neuroimaging data: Whereas early layers are trained globally using standard convolutional layers, we introduce for higher, more abstract layers patch individual filters (PIF). By learning filters in individual image regions (patches) without sharing weights, PIF layers can learn abstract features faster and with fewer samples. We thoroughly evaluated PIF layers for three different tasks and data sets, namely sex classification on UK Biobank data, Alzheimer's disease detection on ADNI data and multiple sclerosis detection on private hospital data. We demonstrate that CNNs using PIF layers result in higher accuracies, especially in low sample size settings, and need fewer training epochs for convergence. To the best of our knowledge, this is the first study which introduces a prior on brain MRI for CNN learning.
Harnessing spatial MRI normalization: patch individual filter layers for CNNs
Nov 14, 2019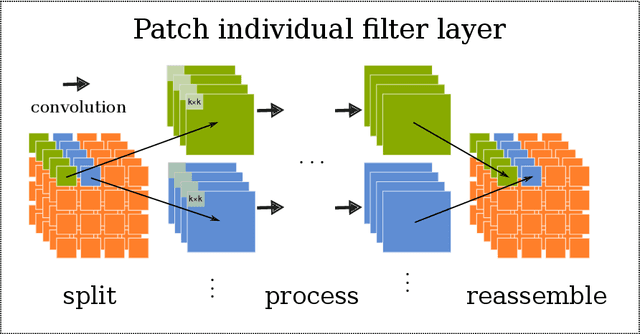
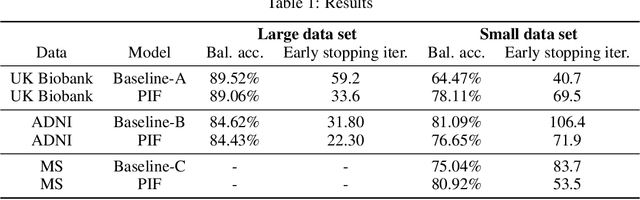
Abstract:Neuroimaging studies based on magnetic resonance imaging (MRI) typically employ rigorous forms of preprocessing. Images are spatially normalized to a standard template using linear and non-linear transformations. Thus, one can assume that a patch at location (x, y, height, width) contains the same brain region across the entire data set. Most analyses applied on brain MRI using convolutional neural networks (CNNs) ignore this distinction from natural images. Here, we suggest a new layer type called patch individual filter (PIF) layer, which trains higher-level filters locally as we assume that more abstract features are locally specific after spatial normalization. We evaluate PIF layers on three different tasks, namely sex classification as well as either Alzheimer's disease (AD) or multiple sclerosis (MS) detection. We demonstrate that CNNs using PIF layers outperform their counterparts in several, especially low sample size settings.
Testing the robustness of attribution methods for convolutional neural networks in MRI-based Alzheimer's disease classification
Sep 19, 2019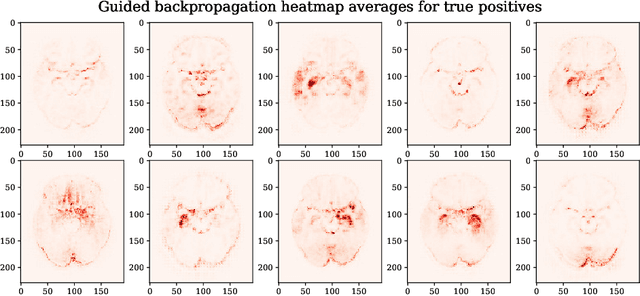
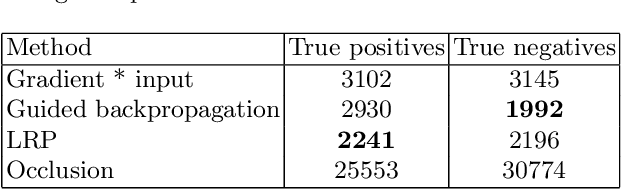
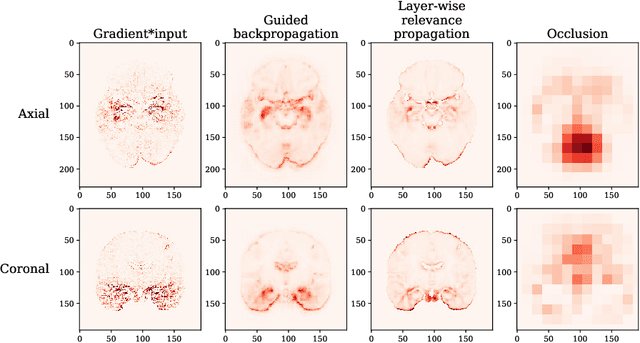
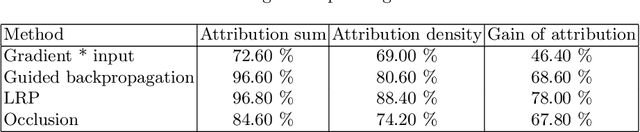
Abstract:Attribution methods are an easy to use tool for investigating and validating machine learning models. Multiple methods have been suggested in the literature and it is not yet clear which method is most suitable for a given task. In this study, we tested the robustness of four attribution methods, namely gradient*input, guided backpropagation, layer-wise relevance propagation and occlusion, for the task of Alzheimer's disease classification. We have repeatedly trained a convolutional neural network (CNN) with identical training settings in order to separate structural MRI data of patients with Alzheimer's disease and healthy controls. Afterwards, we produced attribution maps for each subject in the test data and quantitatively compared them across models and attribution methods. We show that visual comparison is not sufficient and that some widely used attribution methods produce highly inconsistent outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge