Henrik Walter
Automatic rating of incomplete hippocampal inversions evaluated across multiple cohorts
Aug 05, 2024



Abstract:Incomplete Hippocampal Inversion (IHI), sometimes called hippocampal malrotation, is an atypical anatomical pattern of the hippocampus found in about 20% of the general population. IHI can be visually assessed on coronal slices of T1 weighted MR images, using a composite score that combines four anatomical criteria. IHI has been associated with several brain disorders (epilepsy, schizophrenia). However, these studies were based on small samples. Furthermore, the factors (genetic or environmental) that contribute to the genesis of IHI are largely unknown. Large-scale studies are thus needed to further understand IHI and their potential relationships to neurological and psychiatric disorders. However, visual evaluation is long and tedious, justifying the need for an automatic method. In this paper, we propose, for the first time, to automatically rate IHI. We proceed by predicting four anatomical criteria, which are then summed up to form the IHI score, providing the advantage of an interpretable score. We provided an extensive experimental investigation of different machine learning methods and training strategies. We performed automatic rating using a variety of deep learning models (conv5-FC3, ResNet and SECNN) as well as a ridge regression. We studied the generalization of our models using different cohorts and performed multi-cohort learning. We relied on a large population of 2,008 participants from the IMAGEN study, 993 and 403 participants from the QTIM/QTAB studies as well as 985 subjects from the UKBiobank. We showed that deep learning models outperformed a ridge regression. We demonstrated that the performances of the conv5-FC3 network were at least as good as more complex networks while maintaining a low complexity and computation time. We showed that training on a single cohort may lack in variability while training on several cohorts improves generalization.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:016
Promises and pitfalls of deep neural networks in neuroimaging-based psychiatric research
Jan 20, 2023



Abstract:By promising more accurate diagnostics and individual treatment recommendations, deep neural networks and in particular convolutional neural networks have advanced to a powerful tool in medical imaging. Here, we first give an introduction into methodological key concepts and resulting methodological promises including representation and transfer learning, as well as modelling domain-specific priors. After reviewing recent applications within neuroimaging-based psychiatric research, such as the diagnosis of psychiatric diseases, delineation of disease subtypes, normative modeling, and the development of neuroimaging biomarkers, we discuss current challenges. This includes for example the difficulty of training models on small, heterogeneous and biased data sets, the lack of validity of clinical labels, algorithmic bias, and the influence of confounding variables.
Classification of Major Depressive Disorder via Multi-Site Weighted LASSO Model
Jun 03, 2017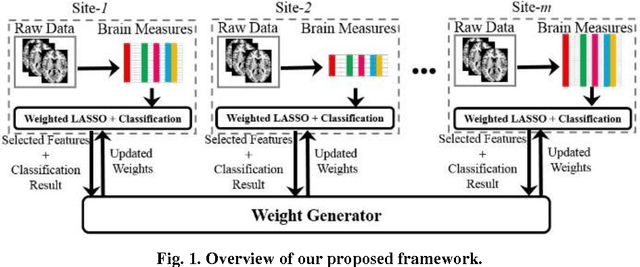
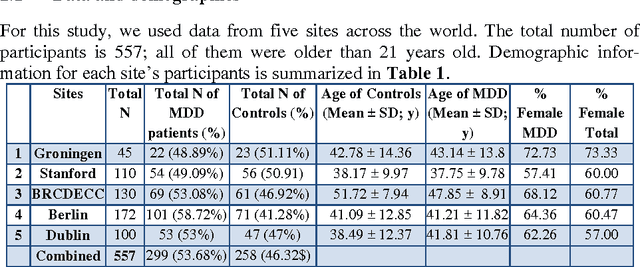
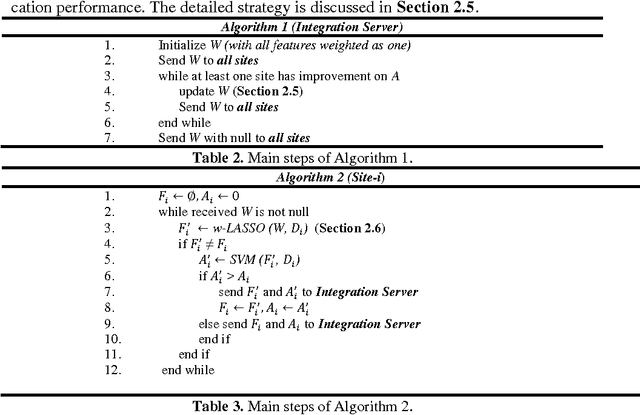
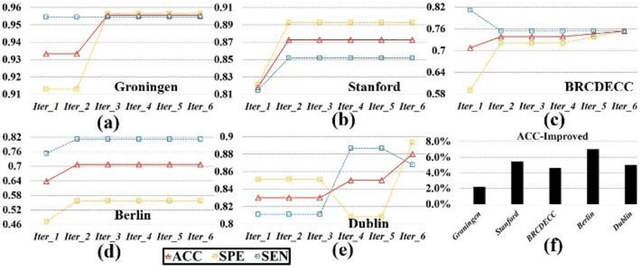
Abstract:Large-scale collaborative analysis of brain imaging data, in psychiatry and neu-rology, offers a new source of statistical power to discover features that boost ac-curacy in disease classification, differential diagnosis, and outcome prediction. However, due to data privacy regulations or limited accessibility to large datasets across the world, it is challenging to efficiently integrate distributed information. Here we propose a novel classification framework through multi-site weighted LASSO: each site performs an iterative weighted LASSO for feature selection separately. Within each iteration, the classification result and the selected features are collected to update the weighting parameters for each feature. This new weight is used to guide the LASSO process at the next iteration. Only the fea-tures that help to improve the classification accuracy are preserved. In tests on da-ta from five sites (299 patients with major depressive disorder (MDD) and 258 normal controls), our method boosted classification accuracy for MDD by 4.9% on average. This result shows the potential of the proposed new strategy as an ef-fective and practical collaborative platform for machine learning on large scale distributed imaging and biobank data.
Nonlinear functional mapping of the human brain
Sep 08, 2015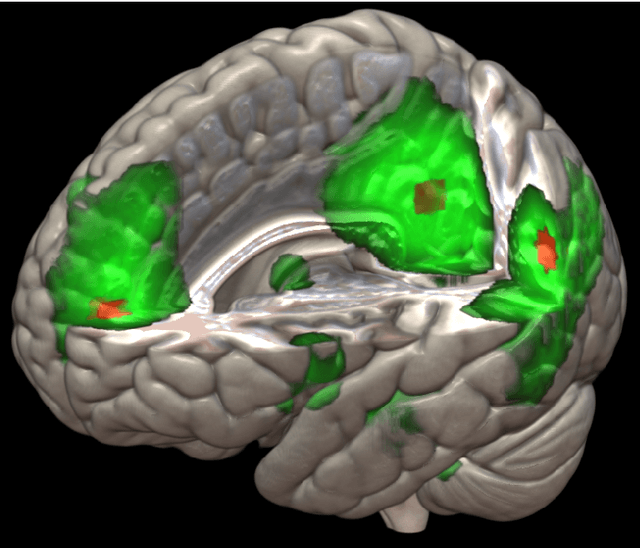
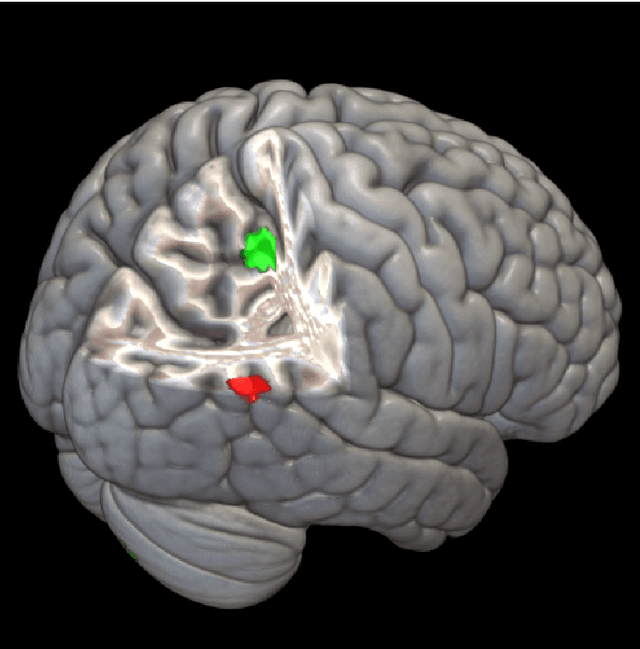
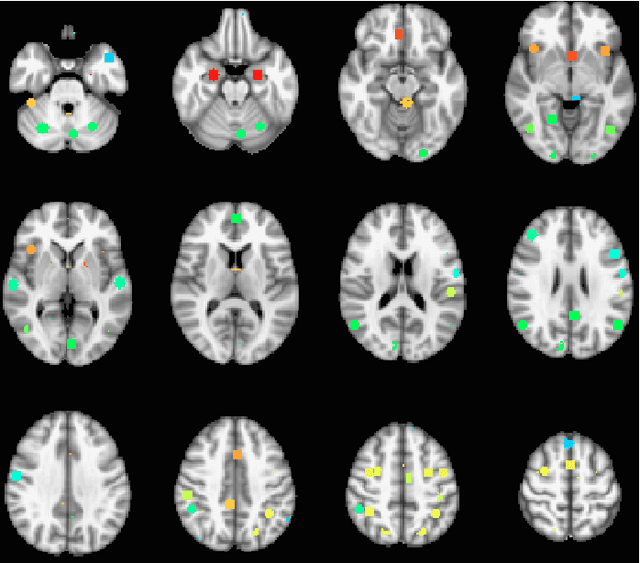
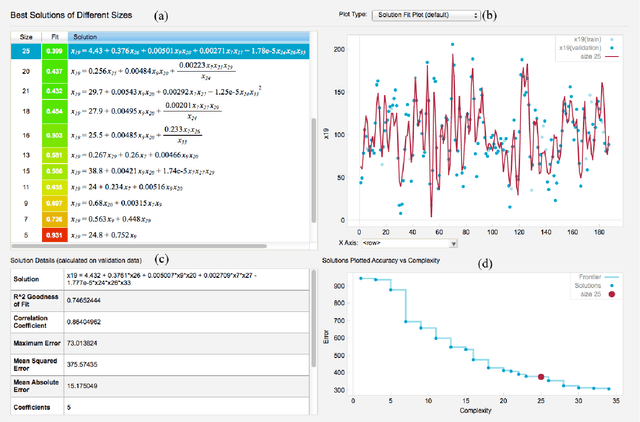
Abstract:The field of neuroimaging has truly become data rich, and novel analytical methods capable of gleaning meaningful information from large stores of imaging data are in high demand. Those methods that might also be applicable on the level of individual subjects, and thus potentially useful clinically, are of special interest. In the present study, we introduce just such a method, called nonlinear functional mapping (NFM), and demonstrate its application in the analysis of resting state fMRI from a 242-subject subset of the IMAGEN project, a European study of adolescents that includes longitudinal phenotypic, behavioral, genetic, and neuroimaging data. NFM employs a computational technique inspired by biological evolution to discover and mathematically characterize interactions among ROI (regions of interest), without making linear or univariate assumptions. We show that statistics of the resulting interaction relationships comport with recent independent work, constituting a preliminary cross-validation. Furthermore, nonlinear terms are ubiquitous in the models generated by NFM, suggesting that some of the interactions characterized here are not discoverable by standard linear methods of analysis. We discuss one such nonlinear interaction in the context of a direct comparison with a procedure involving pairwise correlation, designed to be an analogous linear version of functional mapping. We find another such interaction that suggests a novel distinction in brain function between drinking and non-drinking adolescents: a tighter coupling of ROI associated with emotion, reward, and interoceptive processes such as thirst, among drinkers. Finally, we outline many improvements and extensions of the methodology to reduce computational expense, complement other analytical tools like graph-theoretic analysis, and allow for voxel level NFM to eliminate the necessity of ROI selection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge