Antoine Grigis
Automatic rating of incomplete hippocampal inversions evaluated across multiple cohorts
Aug 05, 2024



Abstract:Incomplete Hippocampal Inversion (IHI), sometimes called hippocampal malrotation, is an atypical anatomical pattern of the hippocampus found in about 20% of the general population. IHI can be visually assessed on coronal slices of T1 weighted MR images, using a composite score that combines four anatomical criteria. IHI has been associated with several brain disorders (epilepsy, schizophrenia). However, these studies were based on small samples. Furthermore, the factors (genetic or environmental) that contribute to the genesis of IHI are largely unknown. Large-scale studies are thus needed to further understand IHI and their potential relationships to neurological and psychiatric disorders. However, visual evaluation is long and tedious, justifying the need for an automatic method. In this paper, we propose, for the first time, to automatically rate IHI. We proceed by predicting four anatomical criteria, which are then summed up to form the IHI score, providing the advantage of an interpretable score. We provided an extensive experimental investigation of different machine learning methods and training strategies. We performed automatic rating using a variety of deep learning models (conv5-FC3, ResNet and SECNN) as well as a ridge regression. We studied the generalization of our models using different cohorts and performed multi-cohort learning. We relied on a large population of 2,008 participants from the IMAGEN study, 993 and 403 participants from the QTIM/QTAB studies as well as 985 subjects from the UKBiobank. We showed that deep learning models outperformed a ridge regression. We demonstrated that the performances of the conv5-FC3 network were at least as good as more complex networks while maintaining a low complexity and computation time. We showed that training on a single cohort may lack in variability while training on several cohorts improves generalization.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:016
Separating common from salient patterns with Contrastive Representation Learning
Feb 19, 2024Abstract:Contrastive Analysis is a sub-field of Representation Learning that aims at separating common factors of variation between two datasets, a background (i.e., healthy subjects) and a target (i.e., diseased subjects), from the salient factors of variation, only present in the target dataset. Despite their relevance, current models based on Variational Auto-Encoders have shown poor performance in learning semantically-expressive representations. On the other hand, Contrastive Representation Learning has shown tremendous performance leaps in various applications (classification, clustering, etc.). In this work, we propose to leverage the ability of Contrastive Learning to learn semantically expressive representations well adapted for Contrastive Analysis. We reformulate it under the lens of the InfoMax Principle and identify two Mutual Information terms to maximize and one to minimize. We decompose the first two terms into an Alignment and a Uniformity term, as commonly done in Contrastive Learning. Then, we motivate a novel Mutual Information minimization strategy to prevent information leakage between common and salient distributions. We validate our method, called SepCLR, on three visual datasets and three medical datasets, specifically conceived to assess the pattern separation capability in Contrastive Analysis. Code available at https://github.com/neurospin-projects/2024_rlouiset_sep_clr.
Can we Agree? On the Rashōmon Effect and the Reliability of Post-Hoc Explainable AI
Aug 14, 2023



Abstract:The Rash\=omon effect poses challenges for deriving reliable knowledge from machine learning models. This study examined the influence of sample size on explanations from models in a Rash\=omon set using SHAP. Experiments on 5 public datasets showed that explanations gradually converged as the sample size increased. Explanations from <128 samples exhibited high variability, limiting reliable knowledge extraction. However, agreement between models improved with more data, allowing for consensus. Bagging ensembles often had higher agreement. The results provide guidance on sufficient data to trust explanations. Variability at low samples suggests that conclusions may be unreliable without validation. Further work is needed with more model types, data domains, and explanation methods. Testing convergence in neural networks and with model-specific explanation methods would be impactful. The approaches explored here point towards principled techniques for eliciting knowledge from ambiguous models.
SepVAE: a contrastive VAE to separate pathological patterns from healthy ones
Jul 12, 2023Abstract:Contrastive Analysis VAE (CA-VAEs) is a family of Variational auto-encoders (VAEs) that aims at separating the common factors of variation between a background dataset (BG) (i.e., healthy subjects) and a target dataset (TG) (i.e., patients) from the ones that only exist in the target dataset. To do so, these methods separate the latent space into a set of salient features (i.e., proper to the target dataset) and a set of common features (i.e., exist in both datasets). Currently, all models fail to prevent the sharing of information between latent spaces effectively and to capture all salient factors of variation. To this end, we introduce two crucial regularization losses: a disentangling term between common and salient representations and a classification term between background and target samples in the salient space. We show a better performance than previous CA-VAEs methods on three medical applications and a natural images dataset (CelebA). Code and datasets are available on GitHub https://github.com/neurospin-projects/2023_rlouiset_sepvae.
Conditional Alignment and Uniformity for Contrastive Learning with Continuous Proxy Labels
Nov 10, 2021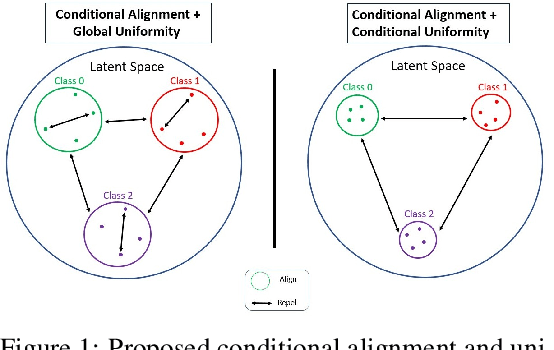
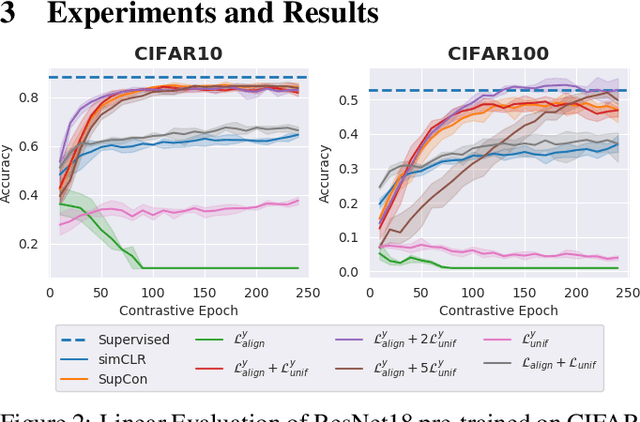
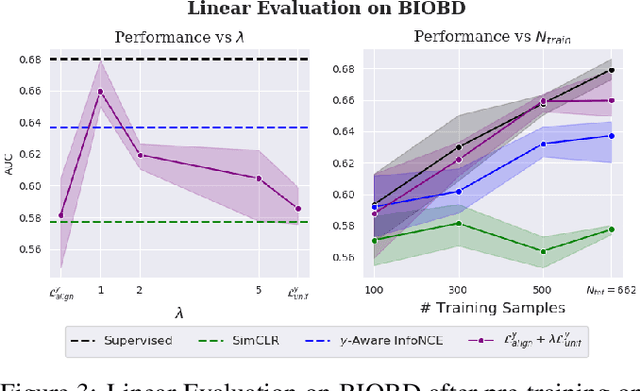
Abstract:Contrastive Learning has shown impressive results on natural and medical images, without requiring annotated data. However, a particularity of medical images is the availability of meta-data (such as age or sex) that can be exploited for learning representations. Here, we show that the recently proposed contrastive y-Aware InfoNCE loss, that integrates multi-dimensional meta-data, asymptotically optimizes two properties: conditional alignment and global uniformity. Similarly to [Wang, 2020], conditional alignment means that similar samples should have similar features, but conditionally on the meta-data. Instead, global uniformity means that the (normalized) features should be uniformly distributed on the unit hyper-sphere, independently of the meta-data. Here, we propose to define conditional uniformity, relying on the meta-data, that repel only samples with dissimilar meta-data. We show that direct optimization of both conditional alignment and uniformity improves the representations, in terms of linear evaluation, on both CIFAR-100 and a brain MRI dataset.
UCSL : A Machine Learning Expectation-Maximization framework for Unsupervised Clustering driven by Supervised Learning
Jul 05, 2021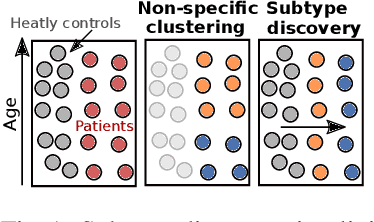
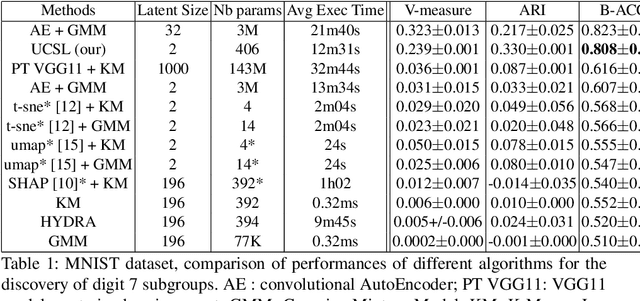
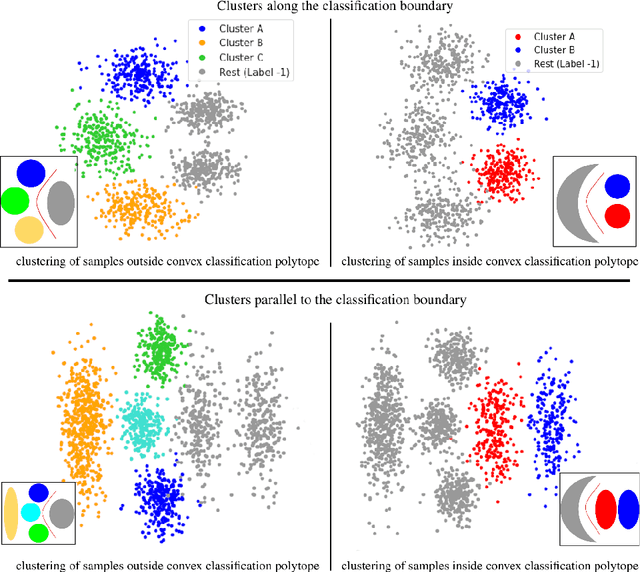
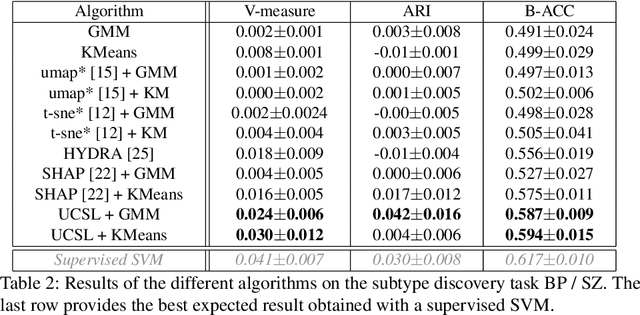
Abstract:Subtype Discovery consists in finding interpretable and consistent sub-parts of a dataset, which are also relevant to a certain supervised task. From a mathematical point of view, this can be defined as a clustering task driven by supervised learning in order to uncover subgroups in line with the supervised prediction. In this paper, we propose a general Expectation-Maximization ensemble framework entitled UCSL (Unsupervised Clustering driven by Supervised Learning). Our method is generic, it can integrate any clustering method and can be driven by both binary classification and regression. We propose to construct a non-linear model by merging multiple linear estimators, one per cluster. Each hyperplane is estimated so that it correctly discriminates - or predict - only one cluster. We use SVC or Logistic Regression for classification and SVR for regression. Furthermore, to perform cluster analysis within a more suitable space, we also propose a dimension-reduction algorithm that projects the data onto an orthonormal space relevant to the supervised task. We analyze the robustness and generalization capability of our algorithm using synthetic and experimental datasets. In particular, we validate its ability to identify suitable consistent sub-types by conducting a psychiatric-diseases cluster analysis with known ground-truth labels. The gain of the proposed method over previous state-of-the-art techniques is about +1.9 points in terms of balanced accuracy. Finally, we make codes and examples available in a scikit-learn-compatible Python package at https://github.com/neurospin-projects/2021_rlouiset_ucsl
* ECML/PKDD 2021
Contrastive Learning with Continuous Proxy Meta-Data for 3D MRI Classification
Jun 16, 2021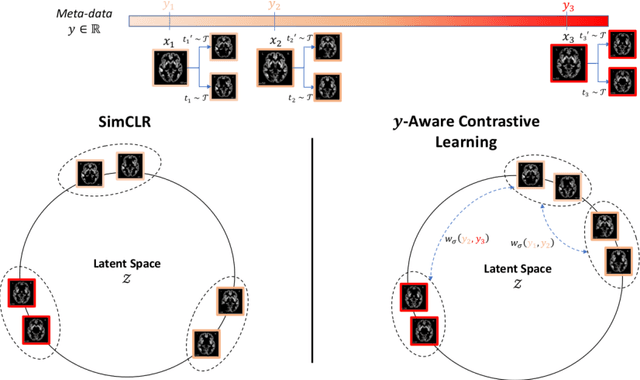
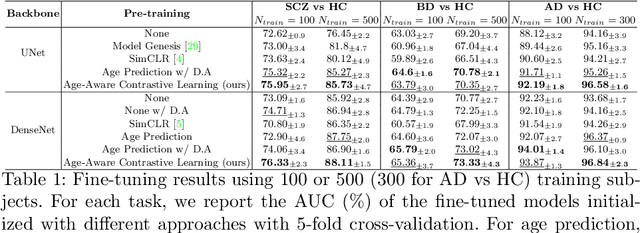
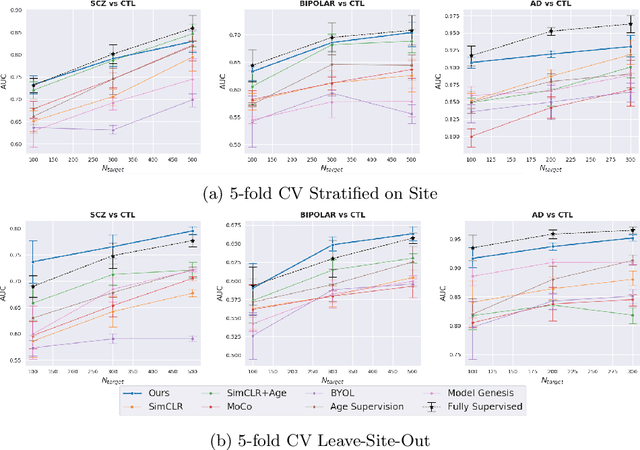
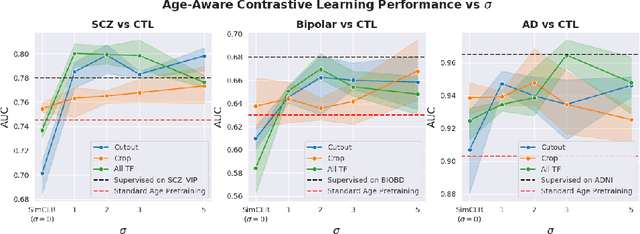
Abstract:Traditional supervised learning with deep neural networks requires a tremendous amount of labelled data to converge to a good solution. For 3D medical images, it is often impractical to build a large homogeneous annotated dataset for a specific pathology. Self-supervised methods offer a new way to learn a representation of the images in an unsupervised manner with a neural network. In particular, contrastive learning has shown great promises by (almost) matching the performance of fully-supervised CNN on vision tasks. Nonetheless, this method does not take advantage of available meta-data, such as participant's age, viewed as prior knowledge. Here, we propose to leverage continuous proxy metadata, in the contrastive learning framework, by introducing a new loss called y-Aware InfoNCE loss. Specifically, we improve the positive sampling during pre-training by adding more positive examples with similar proxy meta-data with the anchor, assuming they share similar discriminative semantic features.With our method, a 3D CNN model pre-trained on $10^4$ multi-site healthy brain MRI scans can extract relevant features for three classification tasks: schizophrenia, bipolar diagnosis and Alzheimer's detection. When fine-tuned, it also outperforms 3D CNN trained from scratch on these tasks, as well as state-of-the-art self-supervised methods. Our code is made publicly available here.
* MICCAI 2021
Benchmarking CNN on 3D Anatomical Brain MRI: Architectures, Data Augmentation and Deep Ensemble Learning
Jun 02, 2021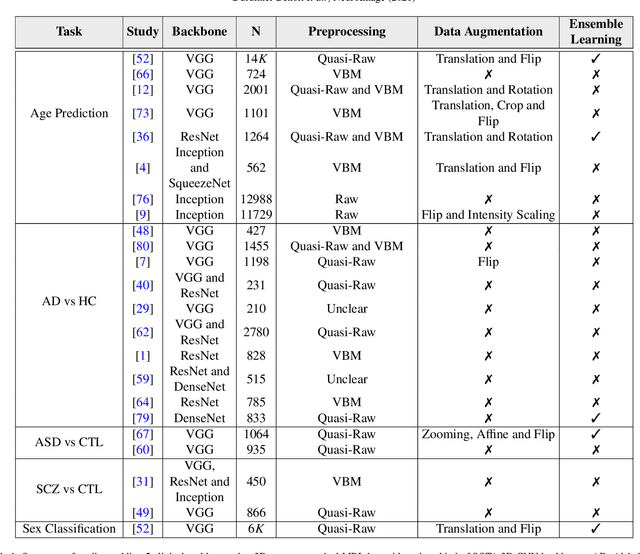
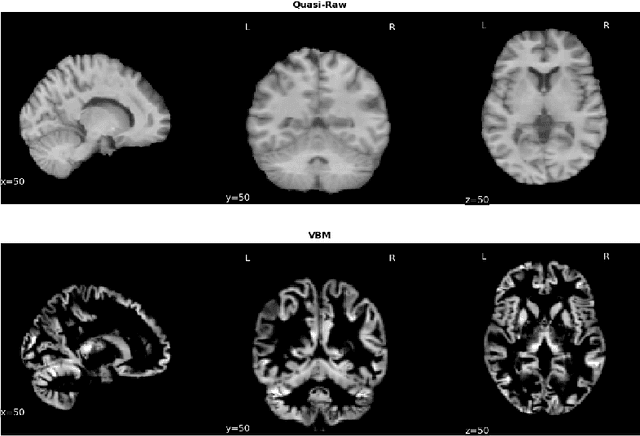
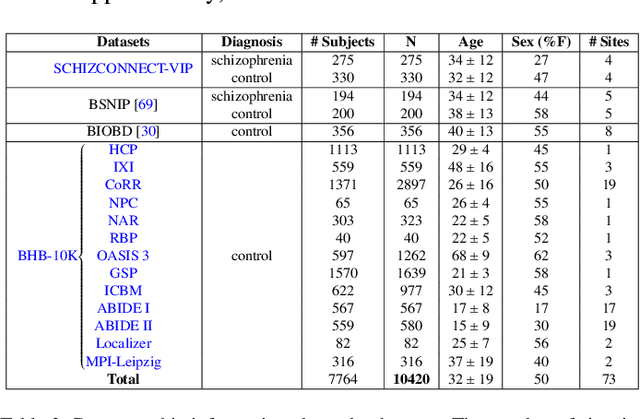
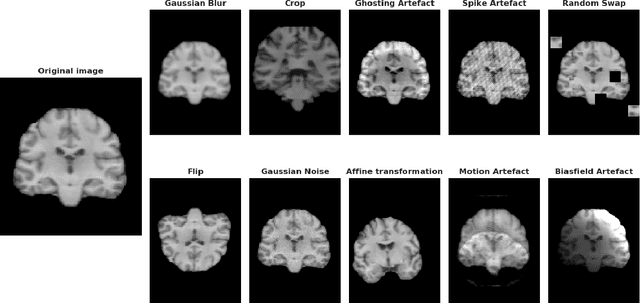
Abstract:Deep Learning (DL) and specifically CNN models have become a de facto method for a wide range of vision tasks, outperforming traditional machine learning (ML) methods. Consequently, they drew a lot of attention in the neuroimaging field in particular for phenotype prediction or computer-aided diagnosis. However, most of the current studies often deal with small single-site cohorts, along with a specific pre-processing pipeline and custom CNN architectures, which make them difficult to compare to. We propose an extensive benchmark of recent state-of-the-art (SOTA) 3D CNN, evaluating also the benefits of data augmentation and deep ensemble learning, on both Voxel-Based Morphometry (VBM) pre-processing and quasi-raw images. Experiments were conducted on a large multi-site 3D brain anatomical MRI data-set comprising N=10k scans on 3 challenging tasks: age prediction, sex classification, and schizophrenia diagnosis. We found that all models provide significantly better predictions with VBM images than quasi-raw data. This finding evolved as the training set approaches 10k samples where quasi-raw data almost reach the performance of VBM. Moreover, we showed that linear models perform comparably with SOTA CNN on VBM data. We also demonstrated that DenseNet and tiny-DenseNet, a lighter version that we proposed, provide a good compromise in terms of performance in all data regime. Therefore, we suggest to employ them as the architectures by default. Critically, we also showed that current CNN are still very biased towards the acquisition site, even when trained with N=10k multi-site images. In this context, VBM pre-processing provides an efficient way to limit this site effect. Surprisingly, we did not find any clear benefit from data augmentation techniques. Finally, we proved that deep ensemble learning is well suited to re-calibrate big CNN models without sacrificing performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge