Robin Louiset
Separating common from salient patterns with Contrastive Representation Learning
Feb 19, 2024Abstract:Contrastive Analysis is a sub-field of Representation Learning that aims at separating common factors of variation between two datasets, a background (i.e., healthy subjects) and a target (i.e., diseased subjects), from the salient factors of variation, only present in the target dataset. Despite their relevance, current models based on Variational Auto-Encoders have shown poor performance in learning semantically-expressive representations. On the other hand, Contrastive Representation Learning has shown tremendous performance leaps in various applications (classification, clustering, etc.). In this work, we propose to leverage the ability of Contrastive Learning to learn semantically expressive representations well adapted for Contrastive Analysis. We reformulate it under the lens of the InfoMax Principle and identify two Mutual Information terms to maximize and one to minimize. We decompose the first two terms into an Alignment and a Uniformity term, as commonly done in Contrastive Learning. Then, we motivate a novel Mutual Information minimization strategy to prevent information leakage between common and salient distributions. We validate our method, called SepCLR, on three visual datasets and three medical datasets, specifically conceived to assess the pattern separation capability in Contrastive Analysis. Code available at https://github.com/neurospin-projects/2024_rlouiset_sep_clr.
Double InfoGAN for Contrastive Analysis
Jan 31, 2024Abstract:Contrastive Analysis (CA) deals with the discovery of what is common and what is distinctive of a target domain compared to a background one. This is of great interest in many applications, such as medical imaging. Current state-of-the-art (SOTA) methods are latent variable models based on VAE (CA-VAEs). However, they all either ignore important constraints or they don't enforce fundamental assumptions. This may lead to sub-optimal solutions where distinctive factors are mistaken for common ones (or viceversa). Furthermore, the generated images have a rather poor quality, typical of VAEs, decreasing their interpretability and usefulness. Here, we propose Double InfoGAN, the first GAN based method for CA that leverages the high-quality synthesis of GAN and the separation power of InfoGAN. Experimental results on four visual datasets, from simple synthetic examples to complex medical images, show that the proposed method outperforms SOTA CA-VAEs in terms of latent separation and image quality. Datasets and code are available online.
SepVAE: a contrastive VAE to separate pathological patterns from healthy ones
Jul 12, 2023Abstract:Contrastive Analysis VAE (CA-VAEs) is a family of Variational auto-encoders (VAEs) that aims at separating the common factors of variation between a background dataset (BG) (i.e., healthy subjects) and a target dataset (TG) (i.e., patients) from the ones that only exist in the target dataset. To do so, these methods separate the latent space into a set of salient features (i.e., proper to the target dataset) and a set of common features (i.e., exist in both datasets). Currently, all models fail to prevent the sharing of information between latent spaces effectively and to capture all salient factors of variation. To this end, we introduce two crucial regularization losses: a disentangling term between common and salient representations and a classification term between background and target samples in the salient space. We show a better performance than previous CA-VAEs methods on three medical applications and a natural images dataset (CelebA). Code and datasets are available on GitHub https://github.com/neurospin-projects/2023_rlouiset_sepvae.
Rethinking Positive Sampling for Contrastive Learning with Kernel
Jun 03, 2022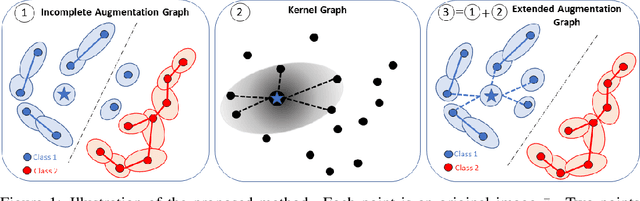
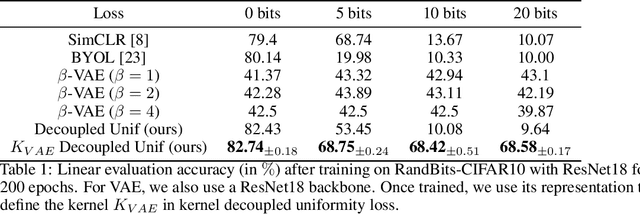
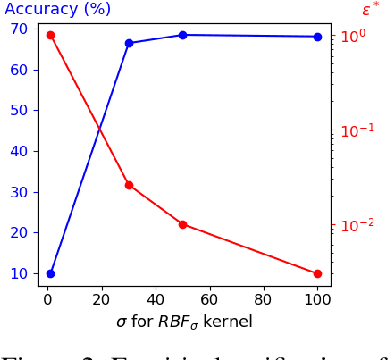
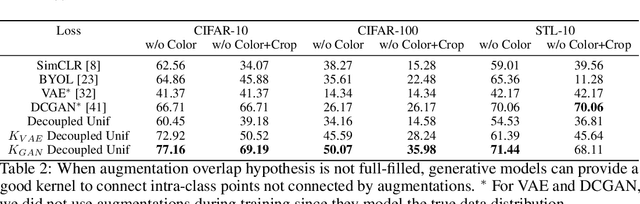
Abstract:Data augmentation is a crucial component in unsupervised contrastive learning (CL). It determines how positive samples are defined and, ultimately, the quality of the representation. While efficient augmentations have been found for standard vision datasets, such as ImageNet, it is still an open problem in other applications, such as medical imaging, or in datasets with easy-to-learn but irrelevant imaging features. In this work, we propose a new way to define positive samples using kernel theory along with a novel loss called decoupled uniformity. We propose to integrate prior information, learnt from generative models or given as auxiliary attributes, into contrastive learning, to make it less dependent on data augmentation. We draw a connection between contrastive learning and the conditional mean embedding theory to derive tight bounds on the downstream classification loss. In an unsupervised setting, we empirically demonstrate that CL benefits from generative models, such as VAE and GAN, to less rely on data augmentations. We validate our framework on vision datasets including CIFAR10, CIFAR100, STL10 and ImageNet100 and a brain MRI dataset. In the weakly supervised setting, we demonstrate that our formulation provides state-of-the-art results.
UCSL : A Machine Learning Expectation-Maximization framework for Unsupervised Clustering driven by Supervised Learning
Jul 05, 2021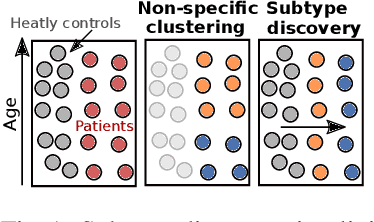
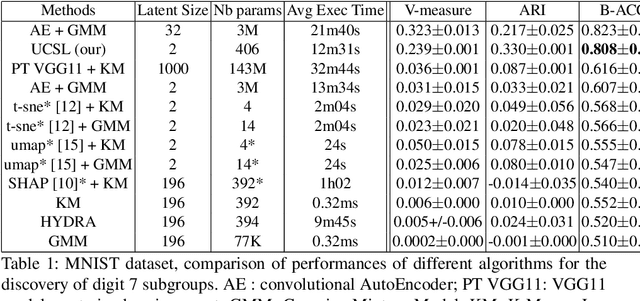
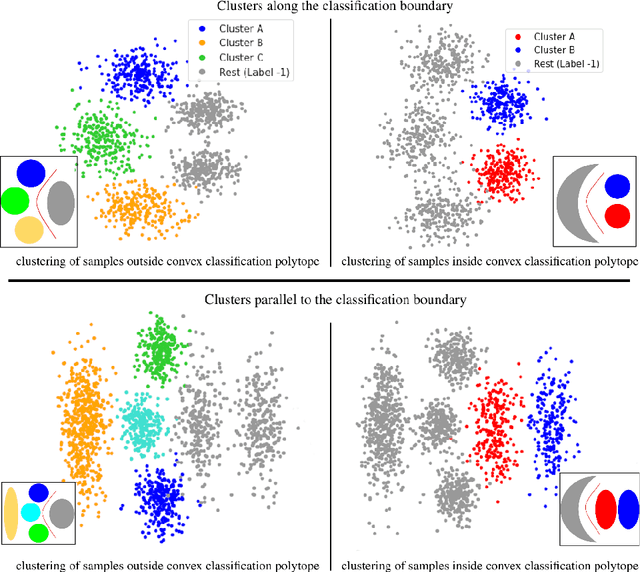
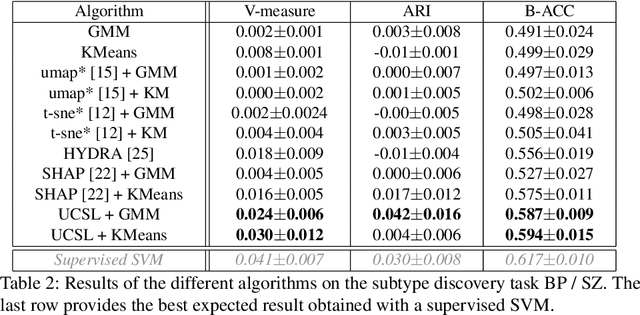
Abstract:Subtype Discovery consists in finding interpretable and consistent sub-parts of a dataset, which are also relevant to a certain supervised task. From a mathematical point of view, this can be defined as a clustering task driven by supervised learning in order to uncover subgroups in line with the supervised prediction. In this paper, we propose a general Expectation-Maximization ensemble framework entitled UCSL (Unsupervised Clustering driven by Supervised Learning). Our method is generic, it can integrate any clustering method and can be driven by both binary classification and regression. We propose to construct a non-linear model by merging multiple linear estimators, one per cluster. Each hyperplane is estimated so that it correctly discriminates - or predict - only one cluster. We use SVC or Logistic Regression for classification and SVR for regression. Furthermore, to perform cluster analysis within a more suitable space, we also propose a dimension-reduction algorithm that projects the data onto an orthonormal space relevant to the supervised task. We analyze the robustness and generalization capability of our algorithm using synthetic and experimental datasets. In particular, we validate its ability to identify suitable consistent sub-types by conducting a psychiatric-diseases cluster analysis with known ground-truth labels. The gain of the proposed method over previous state-of-the-art techniques is about +1.9 points in terms of balanced accuracy. Finally, we make codes and examples available in a scikit-learn-compatible Python package at https://github.com/neurospin-projects/2021_rlouiset_ucsl
* ECML/PKDD 2021
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge