Carlo Alberto Barbano
Robust brain age estimation from structural MRI with contrastive learning
Jul 02, 2025Abstract:Estimating brain age from structural MRI has emerged as a powerful tool for characterizing normative and pathological aging. In this work, we explore contrastive learning as a scalable and robust alternative to supervised approaches for brain age estimation. We introduce a novel contrastive loss function, $\mathcal{L}^{exp}$, and evaluate it across multiple public neuroimaging datasets comprising over 20,000 scans. Our experiments reveal four key findings. First, scaling pre-training on diverse, multi-site data consistently improves generalization performance, cutting external mean absolute error (MAE) nearly in half. Second, $\mathcal{L}^{exp}$ is robust to site-related confounds, maintaining low scanner-predictability as training size increases. Third, contrastive models reliably capture accelerated aging in patients with cognitive impairment and Alzheimer's disease, as shown through brain age gap analysis, ROC curves, and longitudinal trends. Lastly, unlike supervised baselines, $\mathcal{L}^{exp}$ maintains a strong correlation between brain age accuracy and downstream diagnostic performance, supporting its potential as a foundation model for neuroimaging. These results position contrastive learning as a promising direction for building generalizable and clinically meaningful brain representations.
Knowledge Transfer Across Modalities with Natural Language Supervision
Nov 23, 2024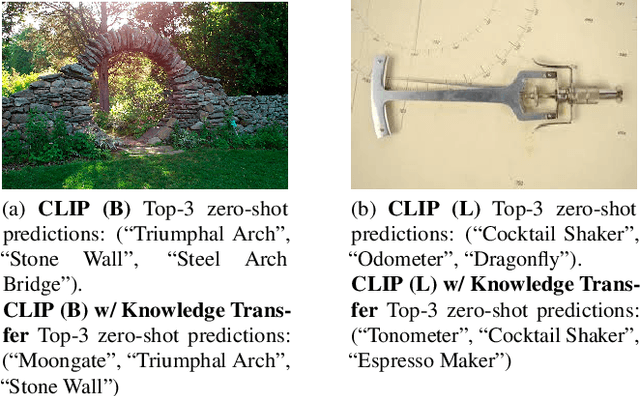
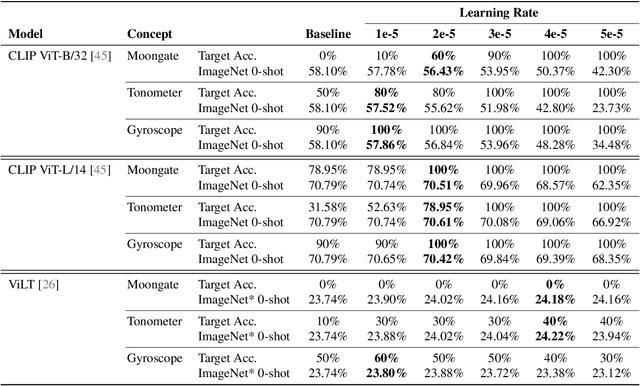
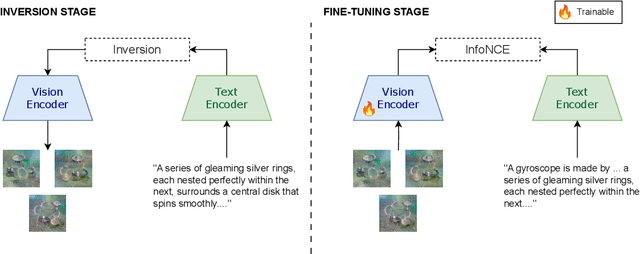
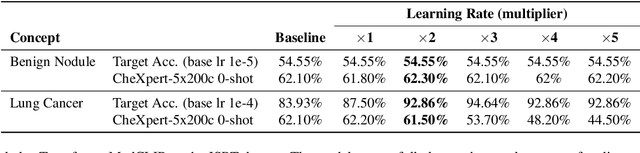
Abstract:We present a way to learn novel concepts by only using their textual description. We call this method Knowledge Transfer. Similarly to human perception, we leverage cross-modal interaction to introduce new concepts. We hypothesize that in a pre-trained visual encoder there are enough low-level features already learned (e.g. shape, appearance, color) that can be used to describe previously unknown high-level concepts. Provided with a textual description of the novel concept, our method works by aligning the known low-level features of the visual encoder to its high-level textual description. We show that Knowledge Transfer can successfully introduce novel concepts in multimodal models, in a very efficient manner, by only requiring a single description of the target concept. Our approach is compatible with both separate textual and visual encoders (e.g. CLIP) and shared parameters across modalities. We also show that, following the same principle, Knowledge Transfer can improve concepts already known by the model. Leveraging Knowledge Transfer we improve zero-shot performance across different tasks such as classification, segmentation, image-text retrieval, and captioning.
Say My Name: a Model's Bias Discovery Framework
Aug 18, 2024Abstract:In the last few years, due to the broad applicability of deep learning to downstream tasks and end-to-end training capabilities, increasingly more concerns about potential biases to specific, non-representative patterns have been raised. Many works focusing on unsupervised debiasing usually leverage the tendency of deep models to learn ``easier'' samples, for example by clustering the latent space to obtain bias pseudo-labels. However, the interpretation of such pseudo-labels is not trivial, especially for a non-expert end user, as it does not provide semantic information about the bias features. To address this issue, we introduce ``Say My Name'' (SaMyNa), the first tool to identify biases within deep models semantically. Unlike existing methods, our approach focuses on biases learned by the model. Our text-based pipeline enhances explainability and supports debiasing efforts: applicable during either training or post-hoc validation, our method can disentangle task-related information and proposes itself as a tool to analyze biases. Evaluation on traditional benchmarks demonstrates its effectiveness in detecting biases and even disclaiming them, showcasing its broad applicability for model diagnosis.
Anatomical Foundation Models for Brain MRIs
Aug 07, 2024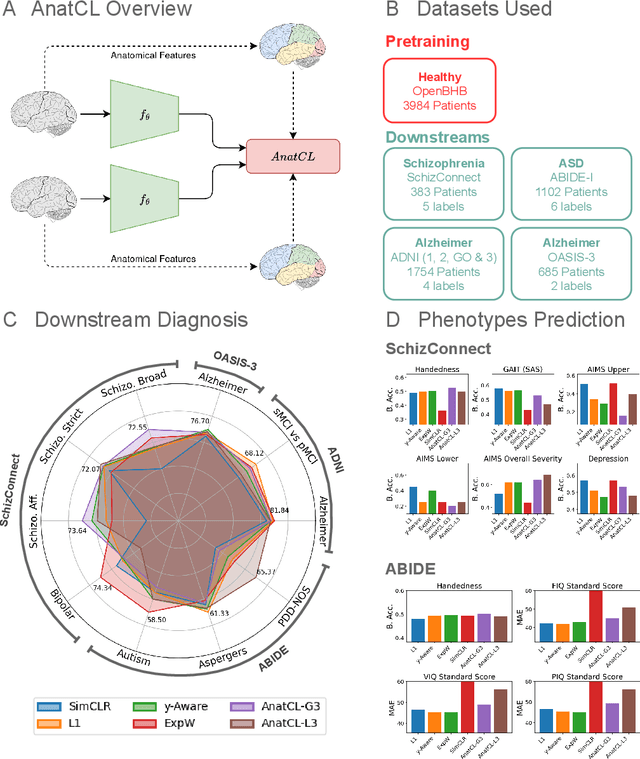
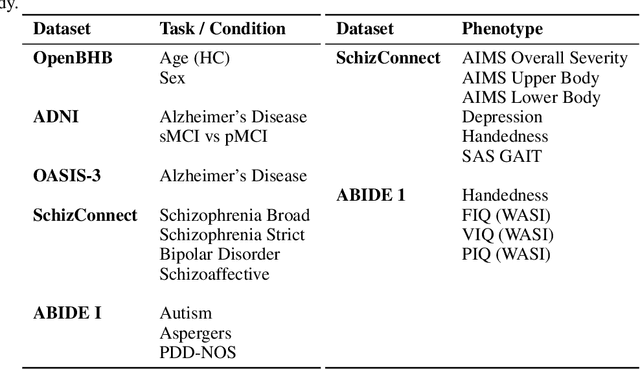
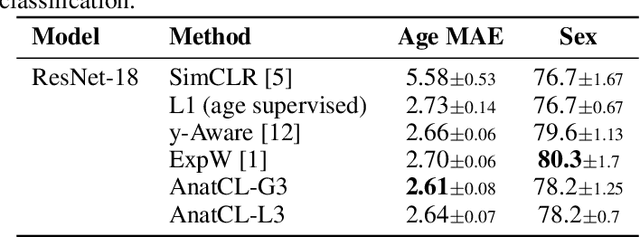
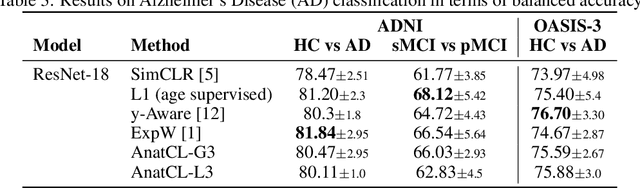
Abstract:Deep Learning (DL) in neuroimaging has become increasingly relevant for detecting neurological conditions and neurodegenerative disorders. One of the most predominant biomarkers in neuroimaging is represented by brain age, which has been shown to be a good indicator for different conditions, such as Alzheimer's Disease. Using brain age for pretraining DL models in transfer learning settings has also recently shown promising results, especially when dealing with data scarcity of different conditions. On the other hand, anatomical information of brain MRIs (e.g. cortical thickness) can provide important information for learning good representations that can be transferred to many downstream tasks. In this work, we propose AnatCL, an anatomical foundation model for brain MRIs that i.) leverages anatomical information with a weakly contrastive learning approach and ii.) achieves state-of-the-art performances in many different downstream tasks. To validate our approach we consider 12 different downstream tasks for diagnosis classification, and prediction of 10 different clinical assessment scores.
Unsupervised Contrastive Analysis for Salient Pattern Detection using Conditional Diffusion Models
Jun 04, 2024



Abstract:Contrastive Analysis (CA) regards the problem of identifying patterns in images that allow distinguishing between a background (BG) dataset (i.e. healthy subjects) and a target (TG) dataset (i.e. unhealthy subjects). Recent works on this topic rely on variational autoencoders (VAE) or contrastive learning strategies to learn the patterns that separate TG samples from BG samples in a supervised manner. However, the dependency on target (unhealthy) samples can be challenging in medical scenarios due to their limited availability. Also, the blurred reconstructions of VAEs lack utility and interpretability. In this work, we redefine the CA task by employing a self-supervised contrastive encoder to learn a latent representation encoding only common patterns from input images, using samples exclusively from the BG dataset during training, and approximating the distribution of the target patterns by leveraging data augmentation techniques. Subsequently, we exploit state-of-the-art generative methods, i.e. diffusion models, conditioned on the learned latent representation to produce a realistic (healthy) version of the input image encoding solely the common patterns. Thorough validation on a facial image dataset and experiments across three brain MRI datasets demonstrate that conditioning the generative process of state-of-the-art generative methods with the latent representation from our self-supervised contrastive encoder yields improvements in the generated image quality and in the accuracy of image classification. The code is available at https://github.com/CristianoPatricio/unsupervised-contrastive-cond-diff.
Multi-target stain normalization for histology slides
Jun 04, 2024


Abstract:Traditional staining normalization approaches, e.g. Macenko, typically rely on the choice of a single representative reference image, which may not adequately account for the diverse staining patterns of datasets collected in practical scenarios. In this study, we introduce a novel approach that leverages multiple reference images to enhance robustness against stain variation. Our method is parameter-free and can be adopted in existing computational pathology pipelines with no significant changes. We evaluate the effectiveness of our method through experiments using a deep-learning pipeline for automatic nuclei segmentation on colorectal images. Our results show that by leveraging multiple reference images, better results can be achieved when generalizing to external data, where the staining can widely differ from the training set.
AI-Assisted Diagnosis for Covid-19 CXR Screening: From Data Collection to Clinical Validation
May 19, 2024



Abstract:In this paper, we present the major results from the Covid Radiographic imaging System based on AI (Co.R.S.A.) project, which took place in Italy. This project aims to develop a state-of-the-art AI-based system for diagnosing Covid-19 pneumonia from Chest X-ray (CXR) images. The contributions of this work are manyfold: the release of the public CORDA dataset, a deep learning pipeline for Covid-19 detection, and the clinical validation of the developed solution by expert radiologists. The proposed detection model is based on a two-step approach that, paired with state-of-the-art debiasing, provides reliable results. Most importantly, our investigation includes the actual usage of the diagnosis aid tool by radiologists, allowing us to assess the real benefits in terms of accuracy and time efficiency. Project homepage: https://corsa.di.unito.it/
Detection of subclinical atherosclerosis by image-based deep learning on chest x-ray
Mar 27, 2024



Abstract:Aims. To develop a deep-learning based system for recognition of subclinical atherosclerosis on a plain frontal chest x-ray. Methods and Results. A deep-learning algorithm to predict coronary artery calcium (CAC) score (the AI-CAC model) was developed on 460 chest x-ray (80% training cohort, 20% internal validation cohort) of primary prevention patients (58.4% male, median age 63 [51-74] years) with available paired chest x-ray and chest computed tomography (CT) indicated for any clinical reason and performed within 3 months. The CAC score calculated on chest CT was used as ground truth. The model was validated on an temporally-independent cohort of 90 patients from the same institution (external validation). The diagnostic accuracy of the AI-CAC model assessed by the area under the curve (AUC) was the primary outcome. Overall, median AI-CAC score was 35 (0-388) and 28.9% patients had no AI-CAC. AUC of the AI-CAC model to identify a CAC>0 was 0.90 in the internal validation cohort and 0.77 in the external validation cohort. Sensitivity was consistently above 92% in both cohorts. In the overall cohort (n=540), among patients with AI-CAC=0, a single ASCVD event occurred, after 4.3 years. Patients with AI-CAC>0 had significantly higher Kaplan Meier estimates for ASCVD events (13.5% vs. 3.4%, log-rank=0.013). Conclusion. The AI-CAC model seems to accurately detect subclinical atherosclerosis on chest x-ray with elevated sensitivity, and to predict ASCVD events with elevated negative predictive value. Adoption of the AI-CAC model to refine CV risk stratification or as an opportunistic screening tool requires prospective evaluation.
Contrastive learning for regression in multi-site brain age prediction
Nov 14, 2022



Abstract:Building accurate Deep Learning (DL) models for brain age prediction is a very relevant topic in neuroimaging, as it could help better understand neurodegenerative disorders and find new biomarkers. To estimate accurate and generalizable models, large datasets have been collected, which are often multi-site and multi-scanner. This large heterogeneity negatively affects the generalization performance of DL models since they are prone to overfit site-related noise. Recently, contrastive learning approaches have been shown to be more robust against noise in data or labels. For this reason, we propose a novel contrastive learning regression loss for robust brain age prediction using MRI scans. Our method achieves state-of-the-art performance on the OpenBHB challenge, yielding the best generalization capability and robustness to site-related noise.
Unbiased Supervised Contrastive Learning
Nov 10, 2022



Abstract:Many datasets are biased, namely they contain easy-to-learn features that are highly correlated with the target class only in the dataset but not in the true underlying distribution of the data. For this reason, learning unbiased models from biased data has become a very relevant research topic in the last years. In this work, we tackle the problem of learning representations that are robust to biases. We first present a margin-based theoretical framework that allows us to clarify why recent contrastive losses (InfoNCE, SupCon, etc.) can fail when dealing with biased data. Based on that, we derive a novel formulation of the supervised contrastive loss (epsilon-SupInfoNCE), providing more accurate control of the minimal distance between positive and negative samples. Furthermore, thanks to our theoretical framework, we also propose FairKL, a new debiasing regularization loss, that works well even with extremely biased data. We validate the proposed losses on standard vision datasets including CIFAR10, CIFAR100, and ImageNet, and we assess the debiasing capability of FairKL with epsilon-SupInfoNCE, reaching state-of-the-art performance on a number of biased datasets, including real instances of biases in the wild.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge