Vito Paolo Pastore
Automated Landmark Detection for assessing hip conditions: A Cross-Modality Validation of MRI versus X-ray
Jan 26, 2026Abstract:Many clinical screening decisions are based on angle measurements. In particular, FemoroAcetabular Impingement (FAI) screening relies on angles traditionally measured on X-rays. However, assessing the height and span of the impingement area requires also a 3D view through an MRI scan. The two modalities inform the surgeon on different aspects of the condition. In this work, we conduct a matched-cohort validation study (89 patients, paired MRI/X-ray) using standard heatmap regression architectures to assess cross-modality clinical equivalence. Seen that landmark detection has been proven effective on X-rays, we show that MRI also achieves equivalent localisation and diagnostic accuracy for cam-type impingement. Our method demonstrates clinical feasibility for FAI assessment in coronal views of 3D MRI volumes, opening the possibility for volumetric analysis through placing further landmarks. These results support integrating automated FAI assessment into routine MRI workflows. Code is released at https://github.com/Malga-Vision/Landmarks-Hip-Conditions
How I Met Your Bias: Investigating Bias Amplification in Diffusion Models
Dec 23, 2025Abstract:Diffusion-based generative models demonstrate state-of-the-art performance across various image synthesis tasks, yet their tendency to replicate and amplify dataset biases remains poorly understood. Although previous research has viewed bias amplification as an inherent characteristic of diffusion models, this work provides the first analysis of how sampling algorithms and their hyperparameters influence bias amplification. We empirically demonstrate that samplers for diffusion models -- commonly optimized for sample quality and speed -- have a significant and measurable effect on bias amplification. Through controlled studies with models trained on Biased MNIST, Multi-Color MNIST and BFFHQ, and with Stable Diffusion, we show that sampling hyperparameters can induce both bias reduction and amplification, even when the trained model is fixed. Source code is available at https://github.com/How-I-met-your-bias/how_i_met_your_bias.
Disentangled representations of microscopy images
Jun 25, 2025Abstract:Microscopy image analysis is fundamental for different applications, from diagnosis to synthetic engineering and environmental monitoring. Modern acquisition systems have granted the possibility to acquire an escalating amount of images, requiring a consequent development of a large collection of deep learning-based automatic image analysis methods. Although deep neural networks have demonstrated great performance in this field, interpretability, an essential requirement for microscopy image analysis, remains an open challenge. This work proposes a Disentangled Representation Learning (DRL) methodology to enhance model interpretability for microscopy image classification. Exploiting benchmark datasets from three different microscopic image domains (plankton, yeast vacuoles, and human cells), we show how a DRL framework, based on transferring a representation learnt from synthetic data, can provide a good trade-off between accuracy and interpretability in this domain.
DIMA: DIffusing Motion Artifacts for unsupervised correction in brain MRI images
Apr 09, 2025



Abstract:Motion artifacts remain a significant challenge in Magnetic Resonance Imaging (MRI), compromising diagnostic quality and potentially leading to misdiagnosis or repeated scans. Existing deep learning approaches for motion artifact correction typically require paired motion-free and motion-affected images for training, which are rarely available in clinical settings. To overcome this requirement, we present DIMA (DIffusing Motion Artifacts), a novel framework that leverages diffusion models to enable unsupervised motion artifact correction in brain MRI. Our two-phase approach first trains a diffusion model on unpaired motion-affected images to learn the distribution of motion artifacts. This model then generates realistic motion artifacts on clean images, creating paired datasets suitable for supervised training of correction networks. Unlike existing methods, DIMA operates without requiring k-space manipulation or detailed knowledge of MRI sequence parameters, making it adaptable across different scanning protocols and hardware. Comprehensive evaluations across multiple datasets and anatomical planes demonstrate that our method achieves comparable performance to state-of-the-art supervised approaches while offering superior generalizability to real clinical data. DIMA represents a significant advancement in making motion artifact correction more accessible for routine clinical use, potentially reducing the need for repeat scans and improving diagnostic accuracy.
Diffusing DeBias: a Recipe for Turning a Bug into a Feature
Feb 13, 2025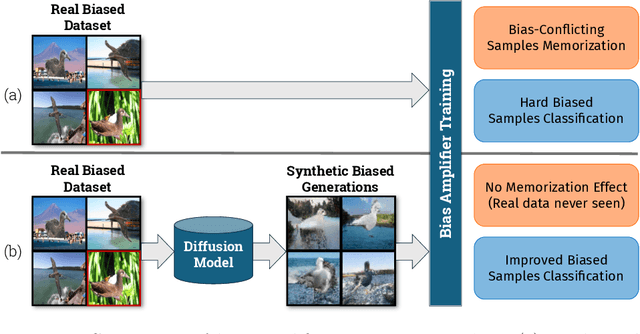
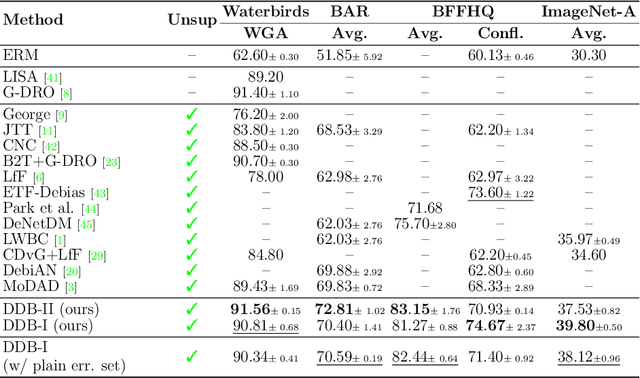
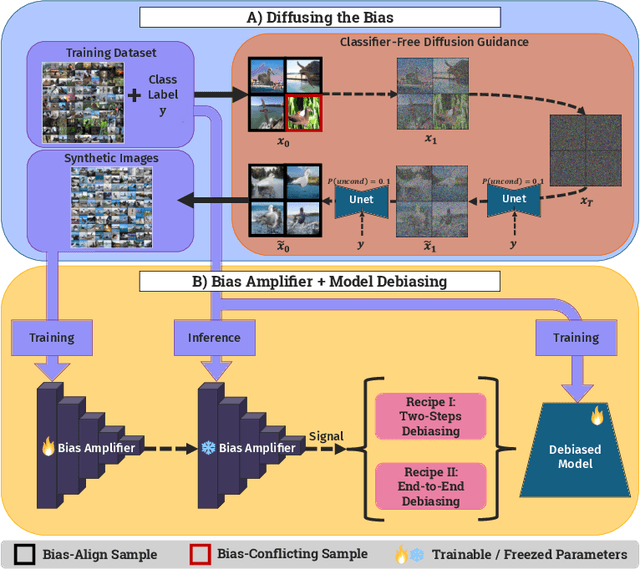

Abstract:Deep learning model effectiveness in classification tasks is often challenged by the quality and quantity of training data which, whenever containing strong spurious correlations between specific attributes and target labels, can result in unrecoverable biases in model predictions. Tackling these biases is crucial in improving model generalization and trust, especially in real-world scenarios. This paper presents Diffusing DeBias (DDB), a novel approach acting as a plug-in for common methods in model debiasing while exploiting the inherent bias-learning tendency of diffusion models. Our approach leverages conditional diffusion models to generate synthetic bias-aligned images, used to train a bias amplifier model, to be further employed as an auxiliary method in different unsupervised debiasing approaches. Our proposed method, which also tackles the common issue of training set memorization typical of this type of tech- niques, beats current state-of-the-art in multiple benchmark datasets by significant margins, demonstrating its potential as a versatile and effective tool for tackling dataset bias in deep learning applications.
Assessing the use of Diffusion models for motion artifact correction in brain MRI
Feb 03, 2025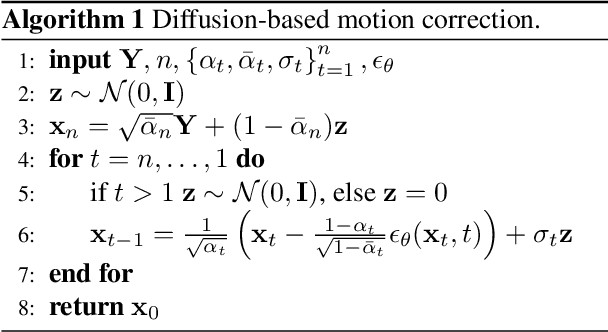
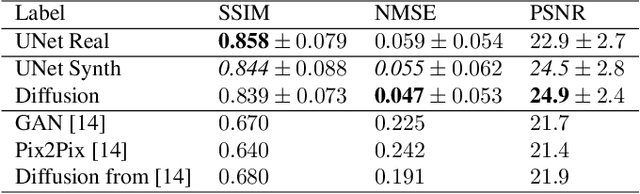
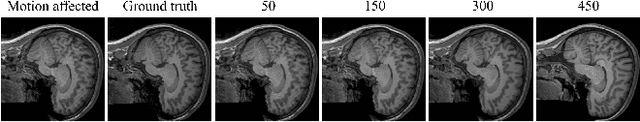
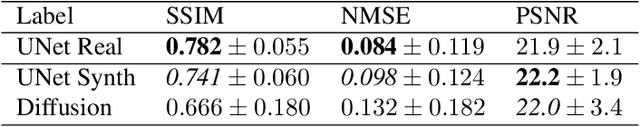
Abstract:Magnetic Resonance Imaging generally requires long exposure times, while being sensitive to patient motion, resulting in artifacts in the acquired images, which may hinder their diagnostic relevance. Despite research efforts to decrease the acquisition time, and designing efficient acquisition sequences, motion artifacts are still a persistent problem, pushing toward the need for the development of automatic motion artifact correction techniques. Recently, diffusion models have been proposed as a solution for the task at hand. While diffusion models can produce high-quality reconstructions, they are also susceptible to hallucination, which poses risks in diagnostic applications. In this study, we critically evaluate the use of diffusion models for correcting motion artifacts in 2D brain MRI scans. Using a popular benchmark dataset, we compare a diffusion model-based approach with state-of-the-art methods consisting of Unets trained in a supervised fashion on motion-affected images to reconstruct ground truth motion-free images. Our findings reveal mixed results: diffusion models can produce accurate predictions or generate harmful hallucinations in this context, depending on data heterogeneity and the acquisition planes considered as input.
Say My Name: a Model's Bias Discovery Framework
Aug 18, 2024Abstract:In the last few years, due to the broad applicability of deep learning to downstream tasks and end-to-end training capabilities, increasingly more concerns about potential biases to specific, non-representative patterns have been raised. Many works focusing on unsupervised debiasing usually leverage the tendency of deep models to learn ``easier'' samples, for example by clustering the latent space to obtain bias pseudo-labels. However, the interpretation of such pseudo-labels is not trivial, especially for a non-expert end user, as it does not provide semantic information about the bias features. To address this issue, we introduce ``Say My Name'' (SaMyNa), the first tool to identify biases within deep models semantically. Unlike existing methods, our approach focuses on biases learned by the model. Our text-based pipeline enhances explainability and supports debiasing efforts: applicable during either training or post-hoc validation, our method can disentangle task-related information and proposes itself as a tool to analyze biases. Evaluation on traditional benchmarks demonstrates its effectiveness in detecting biases and even disclaiming them, showcasing its broad applicability for model diagnosis.
Looking at Model Debiasing through the Lens of Anomaly Detection
Jul 25, 2024


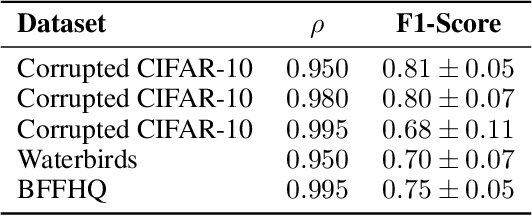
Abstract:It is widely recognized that deep neural networks are sensitive to bias in the data. This means that during training these models are likely to learn spurious correlations between data and labels, resulting in limited generalization abilities and low performance. In this context, model debiasing approaches can be devised aiming at reducing the model's dependency on such unwanted correlations, either leveraging the knowledge of bias information or not. In this work, we focus on the latter and more realistic scenario, showing the importance of accurately predicting the bias-conflicting and bias-aligned samples to obtain compelling performance in bias mitigation. On this ground, we propose to conceive the problem of model bias from an out-of-distribution perspective, introducing a new bias identification method based on anomaly detection. We claim that when data is mostly biased, bias-conflicting samples can be regarded as outliers with respect to the bias-aligned distribution in the feature space of a biased model, thus allowing for precisely detecting them with an anomaly detection method. Coupling the proposed bias identification approach with bias-conflicting data upsampling and augmentation in a two-step strategy, we reach state-of-the-art performance on synthetic and real benchmark datasets. Ultimately, our proposed approach shows that the data bias issue does not necessarily require complex debiasing methods, given that an accurate bias identification procedure is defined.
Self-supervised pre-training with diffusion model for few-shot landmark detection in x-ray images
Jul 25, 2024
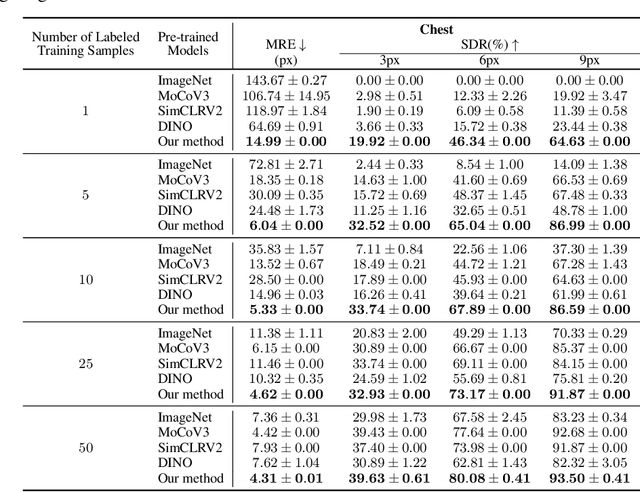


Abstract:In the last few years, deep neural networks have been extensively applied in the medical domain for different tasks, ranging from image classification and segmentation to landmark detection. However, the application of these technologies in the medical domain is often hindered by data scarcity, both in terms of available annotations and images. This study introduces a new self-supervised pre-training protocol based on diffusion models for landmark detection in x-ray images. Our results show that the proposed self-supervised framework can provide accurate landmark detection with a minimal number of available annotated training images (up to 50), outperforming ImageNet supervised pre-training and state-of-the-art self-supervised pre-trainings for three popular x-ray benchmark datasets. To our knowledge, this is the first exploration of diffusion models for self-supervised learning in landmark detection, which may offer a valuable pre-training approach in few-shot regimes, for mitigating data scarcity.
Is in-domain data beneficial in transfer learning for landmarks detection in x-ray images?
Mar 03, 2024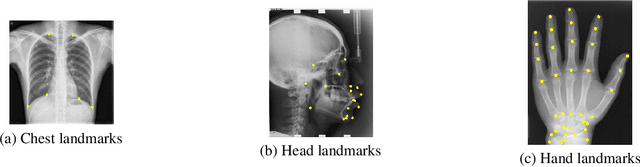
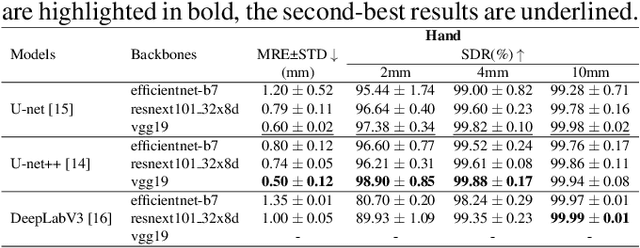
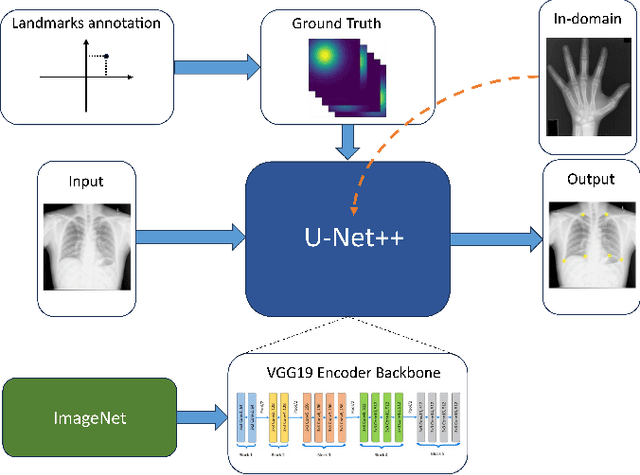
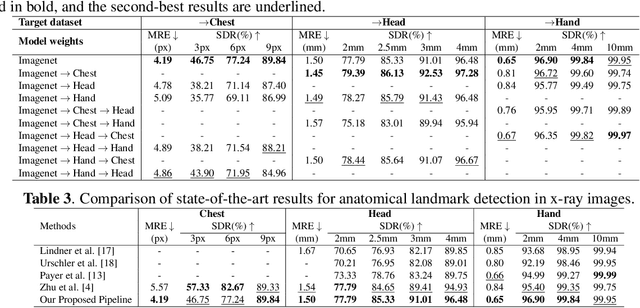
Abstract:In recent years, deep learning has emerged as a promising technique for medical image analysis. However, this application domain is likely to suffer from a limited availability of large public datasets and annotations. A common solution to these challenges in deep learning is the usage of a transfer learning framework, typically with a fine-tuning protocol, where a large-scale source dataset is used to pre-train a model, further fine-tuned on the target dataset. In this paper, we present a systematic study analyzing whether the usage of small-scale in-domain x-ray image datasets may provide any improvement for landmark detection over models pre-trained on large natural image datasets only. We focus on the multi-landmark localization task for three datasets, including chest, head, and hand x-ray images. Our results show that using in-domain source datasets brings marginal or no benefit with respect to an ImageNet out-of-domain pre-training. Our findings can provide an indication for the development of robust landmark detection systems in medical images when no large annotated dataset is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge