Matteo Santacesaria
Deep Unfolding Network for Nonlinear Multi-Frequency Electrical Impedance Tomography
Jul 22, 2025Abstract:Multi-frequency Electrical Impedance Tomography (mfEIT) represents a promising biomedical imaging modality that enables the estimation of tissue conductivities across a range of frequencies. Addressing this challenge, we present a novel variational network, a model-based learning paradigm that strategically merges the advantages and interpretability of classical iterative reconstruction with the power of deep learning. This approach integrates graph neural networks (GNNs) within the iterative Proximal Regularized Gauss Newton (PRGN) framework. By unrolling the PRGN algorithm, where each iteration corresponds to a network layer, we leverage the physical insights of nonlinear model fitting alongside the GNN's capacity to capture inter-frequency correlations. Notably, the GNN architecture preserves the irregular triangular mesh structure used in the solution of the nonlinear forward model, enabling accurate reconstruction of overlapping tissue fraction concentrations.
DIMA: DIffusing Motion Artifacts for unsupervised correction in brain MRI images
Apr 09, 2025



Abstract:Motion artifacts remain a significant challenge in Magnetic Resonance Imaging (MRI), compromising diagnostic quality and potentially leading to misdiagnosis or repeated scans. Existing deep learning approaches for motion artifact correction typically require paired motion-free and motion-affected images for training, which are rarely available in clinical settings. To overcome this requirement, we present DIMA (DIffusing Motion Artifacts), a novel framework that leverages diffusion models to enable unsupervised motion artifact correction in brain MRI. Our two-phase approach first trains a diffusion model on unpaired motion-affected images to learn the distribution of motion artifacts. This model then generates realistic motion artifacts on clean images, creating paired datasets suitable for supervised training of correction networks. Unlike existing methods, DIMA operates without requiring k-space manipulation or detailed knowledge of MRI sequence parameters, making it adaptable across different scanning protocols and hardware. Comprehensive evaluations across multiple datasets and anatomical planes demonstrate that our method achieves comparable performance to state-of-the-art supervised approaches while offering superior generalizability to real clinical data. DIMA represents a significant advancement in making motion artifact correction more accessible for routine clinical use, potentially reducing the need for repeat scans and improving diagnostic accuracy.
TomoSelfDEQ: Self-Supervised Deep Equilibrium Learning for Sparse-Angle CT Reconstruction
Feb 28, 2025Abstract:Deep learning has emerged as a powerful tool for solving inverse problems in imaging, including computed tomography (CT). However, most approaches require paired training data with ground truth images, which can be difficult to obtain, e.g., in medical applications. We present TomoSelfDEQ, a self-supervised Deep Equilibrium (DEQ) framework for sparse-angle CT reconstruction that trains directly on undersampled measurements. We establish theoretical guarantees showing that, under suitable assumptions, our self-supervised updates match those of fully-supervised training with a loss including the (possibly non-unitary) forward operator like the CT forward map. Numerical experiments on sparse-angle CT data confirm this finding, also demonstrating that TomoSelfDEQ outperforms existing self-supervised methods, achieving state-of-the-art results with as few as 16 projection angles.
Assessing the use of Diffusion models for motion artifact correction in brain MRI
Feb 03, 2025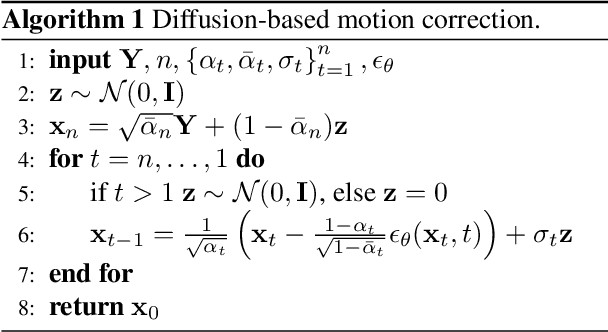
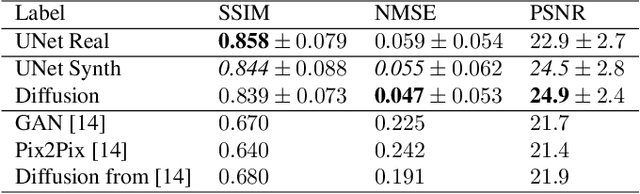
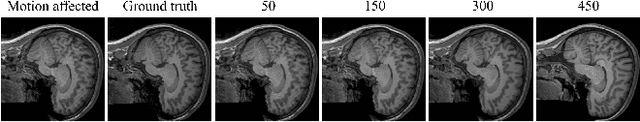
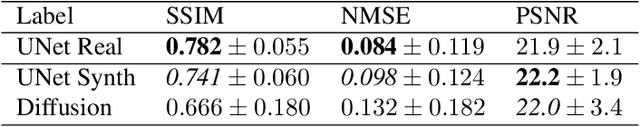
Abstract:Magnetic Resonance Imaging generally requires long exposure times, while being sensitive to patient motion, resulting in artifacts in the acquired images, which may hinder their diagnostic relevance. Despite research efforts to decrease the acquisition time, and designing efficient acquisition sequences, motion artifacts are still a persistent problem, pushing toward the need for the development of automatic motion artifact correction techniques. Recently, diffusion models have been proposed as a solution for the task at hand. While diffusion models can produce high-quality reconstructions, they are also susceptible to hallucination, which poses risks in diagnostic applications. In this study, we critically evaluate the use of diffusion models for correcting motion artifacts in 2D brain MRI scans. Using a popular benchmark dataset, we compare a diffusion model-based approach with state-of-the-art methods consisting of Unets trained in a supervised fashion on motion-affected images to reconstruct ground truth motion-free images. Our findings reveal mixed results: diffusion models can produce accurate predictions or generate harmful hallucinations in this context, depending on data heterogeneity and the acquisition planes considered as input.
Learning sparsity-promoting regularizers for linear inverse problems
Dec 20, 2024Abstract:This paper introduces a novel approach to learning sparsity-promoting regularizers for solving linear inverse problems. We develop a bilevel optimization framework to select an optimal synthesis operator, denoted as $B$, which regularizes the inverse problem while promoting sparsity in the solution. The method leverages statistical properties of the underlying data and incorporates prior knowledge through the choice of $B$. We establish the well-posedness of the optimization problem, provide theoretical guarantees for the learning process, and present sample complexity bounds. The approach is demonstrated through examples, including compact perturbations of a known operator and the problem of learning the mother wavelet, showcasing its flexibility in incorporating prior knowledge into the regularization framework. This work extends previous efforts in Tikhonov regularization by addressing non-differentiable norms and proposing a data-driven approach for sparse regularization in infinite dimensions.
Is in-domain data beneficial in transfer learning for landmarks detection in x-ray images?
Mar 03, 2024
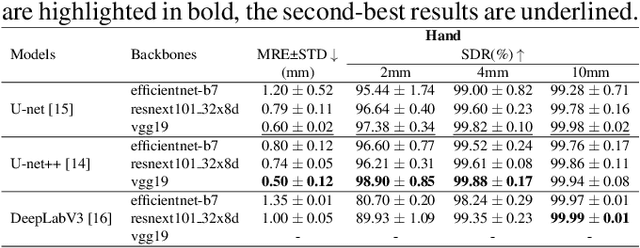
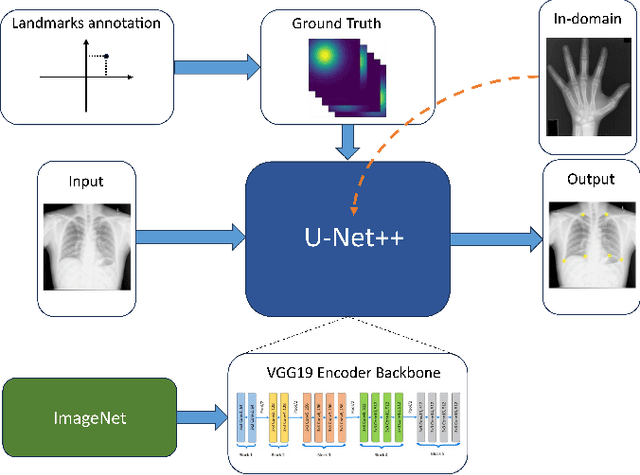
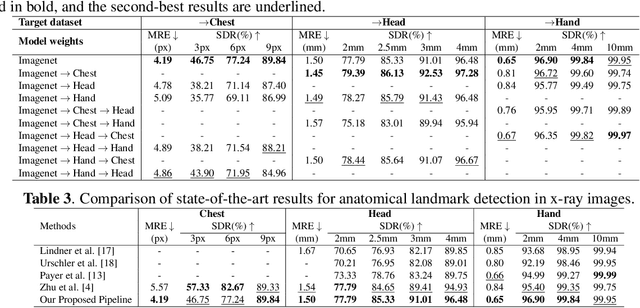
Abstract:In recent years, deep learning has emerged as a promising technique for medical image analysis. However, this application domain is likely to suffer from a limited availability of large public datasets and annotations. A common solution to these challenges in deep learning is the usage of a transfer learning framework, typically with a fine-tuning protocol, where a large-scale source dataset is used to pre-train a model, further fine-tuned on the target dataset. In this paper, we present a systematic study analyzing whether the usage of small-scale in-domain x-ray image datasets may provide any improvement for landmark detection over models pre-trained on large natural image datasets only. We focus on the multi-landmark localization task for three datasets, including chest, head, and hand x-ray images. Our results show that using in-domain source datasets brings marginal or no benefit with respect to an ImageNet out-of-domain pre-training. Our findings can provide an indication for the development of robust landmark detection systems in medical images when no large annotated dataset is available.
Learning a Gaussian Mixture for Sparsity Regularization in Inverse Problems
Jan 29, 2024



Abstract:In inverse problems, it is widely recognized that the incorporation of a sparsity prior yields a regularization effect on the solution. This approach is grounded on the a priori assumption that the unknown can be appropriately represented in a basis with a limited number of significant components, while most coefficients are close to zero. This occurrence is frequently observed in real-world scenarios, such as with piecewise smooth signals. In this study, we propose a probabilistic sparsity prior formulated as a mixture of degenerate Gaussians, capable of modeling sparsity with respect to a generic basis. Under this premise, we design a neural network that can be interpreted as the Bayes estimator for linear inverse problems. Additionally, we put forth both a supervised and an unsupervised training strategy to estimate the parameters of this network. To evaluate the effectiveness of our approach, we conduct a numerical comparison with commonly employed sparsity-promoting regularization techniques, namely LASSO, group LASSO, iterative hard thresholding, and sparse coding/dictionary learning. Notably, our reconstructions consistently exhibit lower mean square error values across all $1$D datasets utilized for the comparisons, even in cases where the datasets significantly deviate from a Gaussian mixture model.
Manifold Learning by Mixture Models of VAEs for Inverse Problems
Mar 27, 2023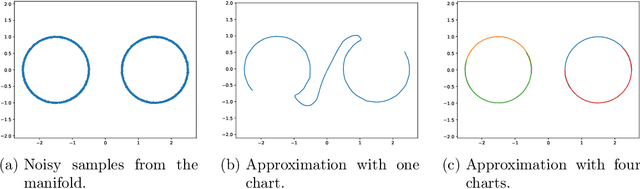
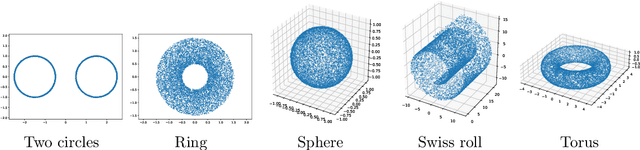
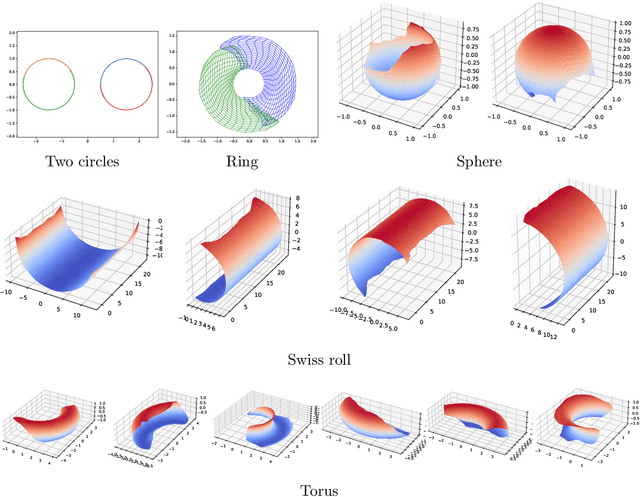
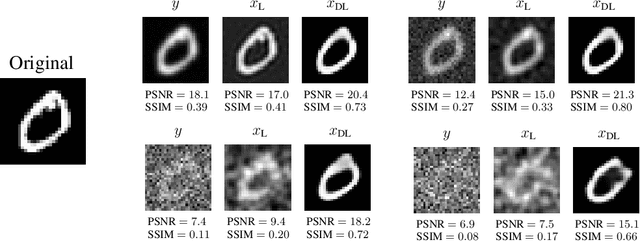
Abstract:Representing a manifold of very high-dimensional data with generative models has been shown to be computationally efficient in practice. However, this requires that the data manifold admits a global parameterization. In order to represent manifolds of arbitrary topology, we propose to learn a mixture model of variational autoencoders. Here, every encoder-decoder pair represents one chart of a manifold. We propose a loss function for maximum likelihood estimation of the model weights and choose an architecture that provides us the analytical expression of the charts and of their inverses. Once the manifold is learned, we use it for solving inverse problems by minimizing a data fidelity term restricted to the learned manifold. To solve the arising minimization problem we propose a Riemannian gradient descent algorithm on the learned manifold. We demonstrate the performance of our method for low-dimensional toy examples as well as for deblurring and electrical impedance tomography on certain image manifolds.
Continuous Generative Neural Networks
May 29, 2022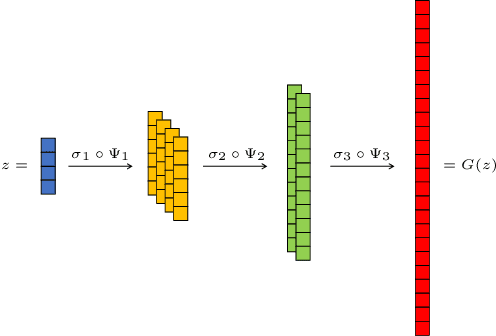
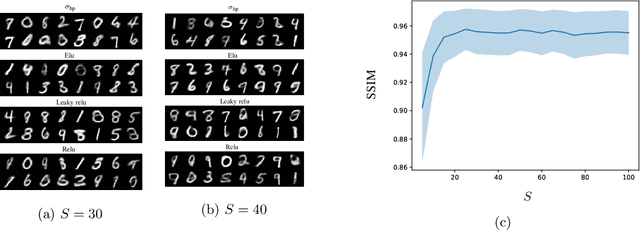
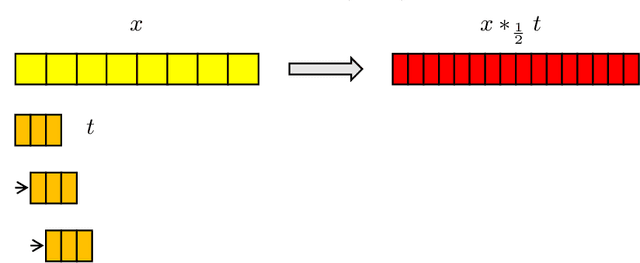
Abstract:In this work, we present and study Continuous Generative Neural Networks (CGNNs), namely, generative models in the continuous setting. The architecture is inspired by DCGAN, with one fully connected layer, several convolutional layers and nonlinear activation functions. In the continuous $L^2$ setting, the dimensions of the spaces of each layer are replaced by the scales of a multiresolution analysis of a compactly supported wavelet. We present conditions on the convolutional filters and on the nonlinearity that guarantee that a CGNN is injective. This theory finds applications to inverse problems, and allows for deriving Lipschitz stability estimates for (possibly nonlinear) infinite-dimensional inverse problems with unknowns belonging to the manifold generated by a CGNN. Several numerical simulations, including image deblurring, illustrate and validate this approach.
Learning the optimal regularizer for inverse problems
Jun 11, 2021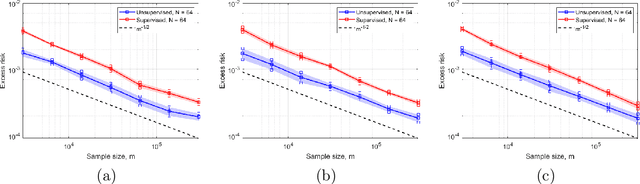
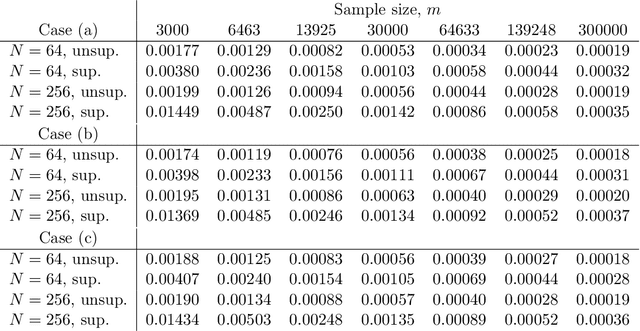
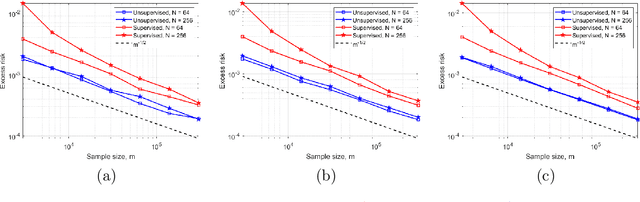
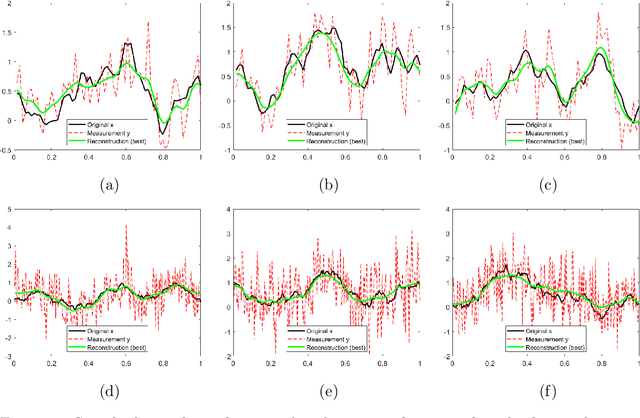
Abstract:In this work, we consider the linear inverse problem $y=Ax+\epsilon$, where $A\colon X\to Y$ is a known linear operator between the separable Hilbert spaces $X$ and $Y$, $x$ is a random variable in $X$ and $\epsilon$ is a zero-mean random process in $Y$. This setting covers several inverse problems in imaging including denoising, deblurring, and X-ray tomography. Within the classical framework of regularization, we focus on the case where the regularization functional is not given a priori but learned from data. Our first result is a characterization of the optimal generalized Tikhonov regularizer, with respect to the mean squared error. We find that it is completely independent of the forward operator $A$ and depends only on the mean and covariance of $x$. Then, we consider the problem of learning the regularizer from a finite training set in two different frameworks: one supervised, based on samples of both $x$ and $y$, and one unsupervised, based only on samples of $x$. In both cases, we prove generalization bounds, under some weak assumptions on the distribution of $x$ and $\epsilon$, including the case of sub-Gaussian variables. Our bounds hold in infinite-dimensional spaces, thereby showing that finer and finer discretizations do not make this learning problem harder. The results are validated through numerical simulations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge