James H. Cole
A self-supervised text-vision framework for automated brain abnormality detection
May 05, 2024



Abstract:Artificial neural networks trained on large, expert-labelled datasets are considered state-of-the-art for a range of medical image recognition tasks. However, categorically labelled datasets are time-consuming to generate and constrain classification to a pre-defined, fixed set of classes. For neuroradiological applications in particular, this represents a barrier to clinical adoption. To address these challenges, we present a self-supervised text-vision framework that learns to detect clinically relevant abnormalities in brain MRI scans by directly leveraging the rich information contained in accompanying free-text neuroradiology reports. Our training approach consisted of two-steps. First, a dedicated neuroradiological language model - NeuroBERT - was trained to generate fixed-dimensional vector representations of neuroradiology reports (N = 50,523) via domain-specific self-supervised learning tasks. Next, convolutional neural networks (one per MRI sequence) learnt to map individual brain scans to their corresponding text vector representations by optimising a mean square error loss. Once trained, our text-vision framework can be used to detect abnormalities in unreported brain MRI examinations by scoring scans against suitable query sentences (e.g., 'there is an acute stroke', 'there is hydrocephalus' etc.), enabling a range of classification-based applications including automated triage. Potentially, our framework could also serve as a clinical decision support tool, not only by suggesting findings to radiologists and detecting errors in provisional reports, but also by retrieving and displaying examples of pathologies from historical examinations that could be relevant to the current case based on textual descriptors.
Distributional Gaussian Processes Layers for Out-of-Distribution Detection
Jun 27, 2022


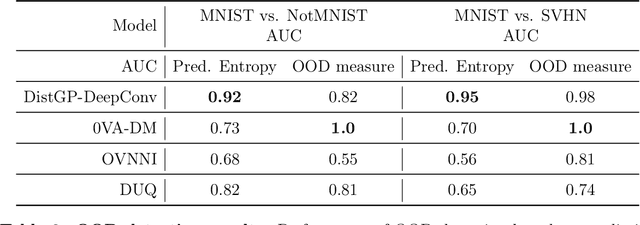
Abstract:Machine learning models deployed on medical imaging tasks must be equipped with out-of-distribution detection capabilities in order to avoid erroneous predictions. It is unsure whether out-of-distribution detection models reliant on deep neural networks are suitable for detecting domain shifts in medical imaging. Gaussian Processes can reliably separate in-distribution data points from out-of-distribution data points via their mathematical construction. Hence, we propose a parameter efficient Bayesian layer for hierarchical convolutional Gaussian Processes that incorporates Gaussian Processes operating in Wasserstein-2 space to reliably propagate uncertainty. This directly replaces convolving Gaussian Processes with a distance-preserving affine operator on distributions. Our experiments on brain tissue-segmentation show that the resulting architecture approaches the performance of well-established deterministic segmentation algorithms (U-Net), which has not been achieved with previous hierarchical Gaussian Processes. Moreover, by applying the same segmentation model to out-of-distribution data (i.e., images with pathology such as brain tumors), we show that our uncertainty estimates result in out-of-distribution detection that outperforms the capabilities of previous Bayesian networks and reconstruction-based approaches that learn normative distributions. To facilitate future work our code is publicly available.
An Uncertainty-Aware, Shareable and Transparent Neural Network Architecture for Brain-Age Modeling
Jul 16, 2021
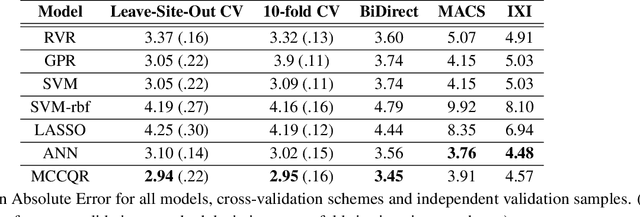
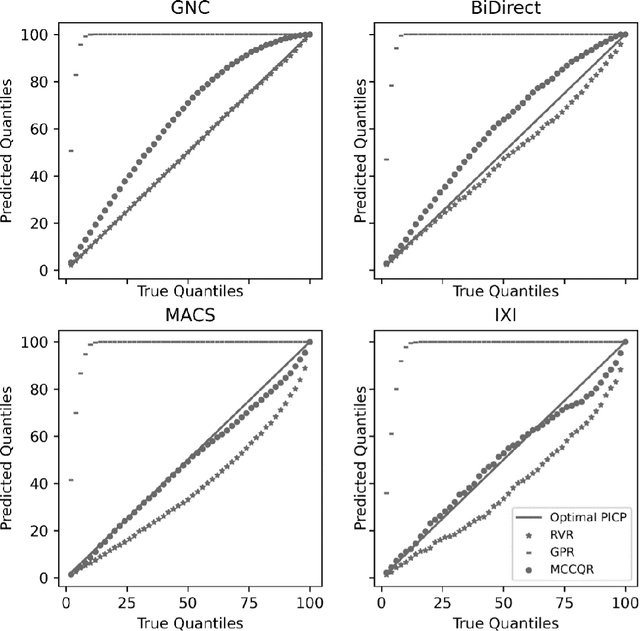
Abstract:The deviation between chronological age and age predicted from neuroimaging data has been identified as a sensitive risk-marker of cross-disorder brain changes, growing into a cornerstone of biological age-research. However, Machine Learning models underlying the field do not consider uncertainty, thereby confounding results with training data density and variability. Also, existing models are commonly based on homogeneous training sets, often not independently validated, and cannot be shared due to data protection issues. Here, we introduce an uncertainty-aware, shareable, and transparent Monte-Carlo Dropout Composite-Quantile-Regression (MCCQR) Neural Network trained on N=10,691 datasets from the German National Cohort. The MCCQR model provides robust, distribution-free uncertainty quantification in high-dimensional neuroimaging data, achieving lower error rates compared to existing models across ten recruitment centers and in three independent validation samples (N=4,004). In two examples, we demonstrate that it prevents spurious associations and increases power to detect accelerated brain-aging. We make the pre-trained model publicly available.
Automated triaging of head MRI examinations using convolutional neural networks
Jun 15, 2021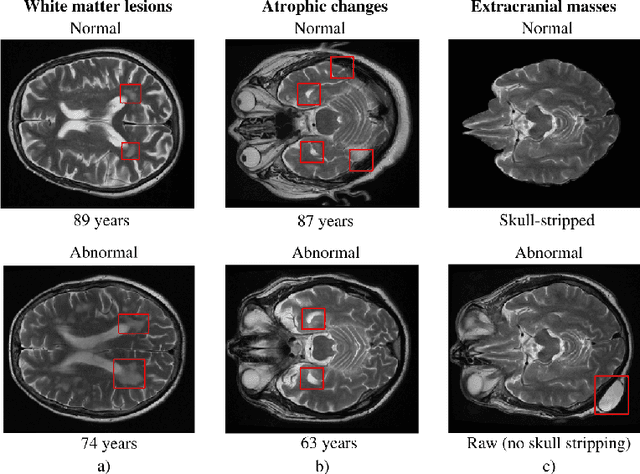
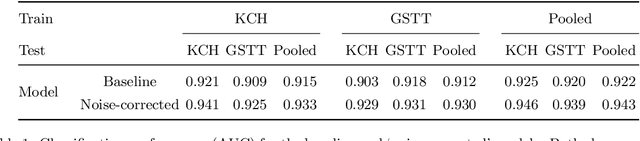
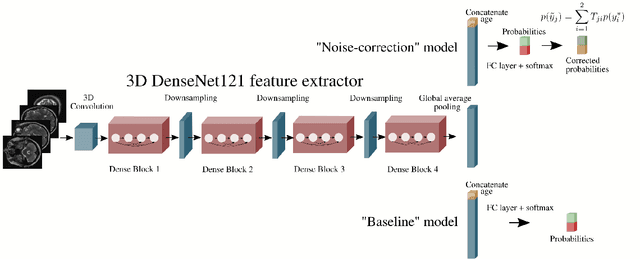
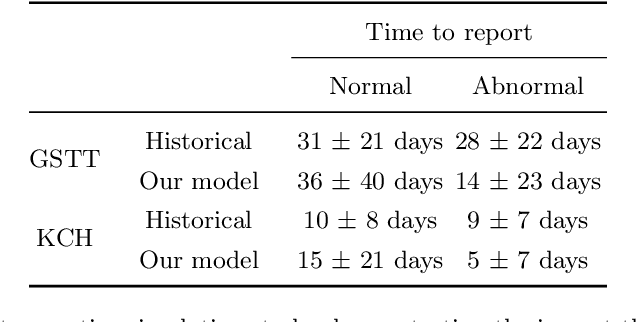
Abstract:The growing demand for head magnetic resonance imaging (MRI) examinations, along with a global shortage of radiologists, has led to an increase in the time taken to report head MRI scans around the world. For many neurological conditions, this delay can result in increased morbidity and mortality. An automated triaging tool could reduce reporting times for abnormal examinations by identifying abnormalities at the time of imaging and prioritizing the reporting of these scans. In this work, we present a convolutional neural network for detecting clinically-relevant abnormalities in $\text{T}_2$-weighted head MRI scans. Using a validated neuroradiology report classifier, we generated a labelled dataset of 43,754 scans from two large UK hospitals for model training, and demonstrate accurate classification (area under the receiver operating curve (AUC) = 0.943) on a test set of 800 scans labelled by a team of neuroradiologists. Importantly, when trained on scans from only a single hospital the model generalized to scans from the other hospital ($\Delta$AUC $\leq$ 0.02). A simulation study demonstrated that our model would reduce the mean reporting time for abnormal examinations from 28 days to 14 days and from 9 days to 5 days at the two hospitals, demonstrating feasibility for use in a clinical triage environment.
Distributional Gaussian Process Layers for Outlier Detection in Image Segmentation
Apr 28, 2021
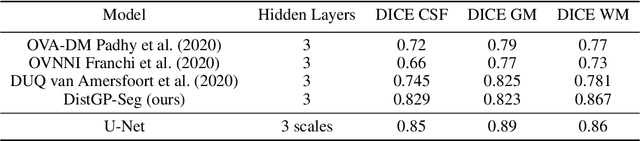
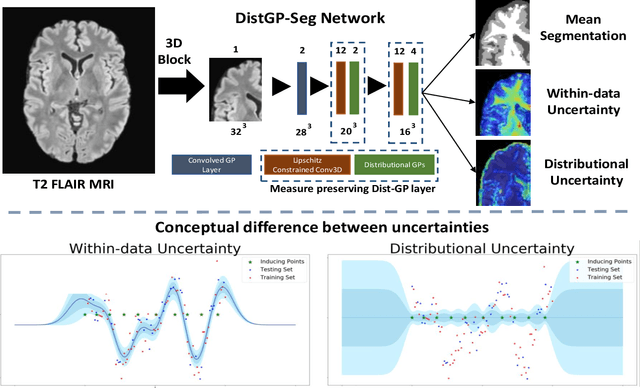
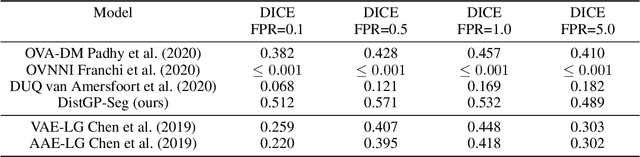
Abstract:We propose a parameter efficient Bayesian layer for hierarchical convolutional Gaussian Processes that incorporates Gaussian Processes operating in Wasserstein-2 space to reliably propagate uncertainty. This directly replaces convolving Gaussian Processes with a distance-preserving affine operator on distributions. Our experiments on brain tissue-segmentation show that the resulting architecture approaches the performance of well-established deterministic segmentation algorithms (U-Net), which has never been achieved with previous hierarchical Gaussian Processes. Moreover, by applying the same segmentation model to out-of-distribution data (i.e., images with pathology such as brain tumors), we show that our uncertainty estimates result in out-of-distribution detection that outperforms the capabilities of previous Bayesian networks and reconstruction-based approaches that learn normative distributions.
Labelling imaging datasets on the basis of neuroradiology reports: a validation study
Jul 08, 2020

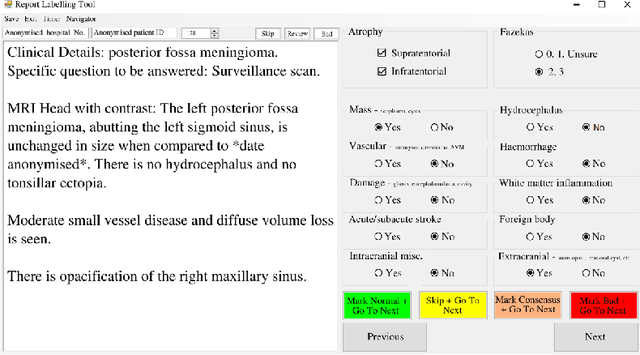
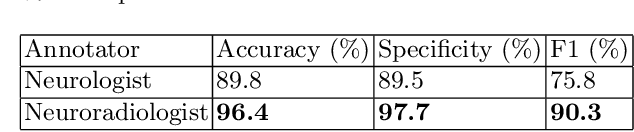
Abstract:Natural language processing (NLP) shows promise as a means to automate the labelling of hospital-scale neuroradiology magnetic resonance imaging (MRI) datasets for computer vision applications. To date, however, there has been no thorough investigation into the validity of this approach, including determining the accuracy of report labels compared to image labels as well as examining the performance of non-specialist labellers. In this work, we draw on the experience of a team of neuroradiologists who labelled over 5000 MRI neuroradiology reports as part of a project to build a dedicated deep learning-based neuroradiology report classifier. We show that, in our experience, assigning binary labels (i.e. normal vs abnormal) to images from reports alone is highly accurate. In contrast to the binary labels, however, the accuracy of more granular labelling is dependent on the category, and we highlight reasons for this discrepancy. We also show that downstream model performance is reduced when labelling of training reports is performed by a non-specialist. To allow other researchers to accelerate their research, we make our refined abnormality definitions and labelling rules available, as well as our easy-to-use radiology report labelling app which helps streamline this process.
Automated Labelling using an Attention model for Radiology reports of MRI scans (ALARM)
Feb 16, 2020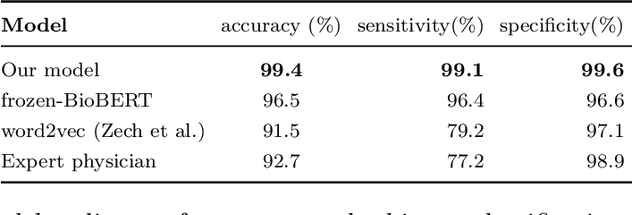
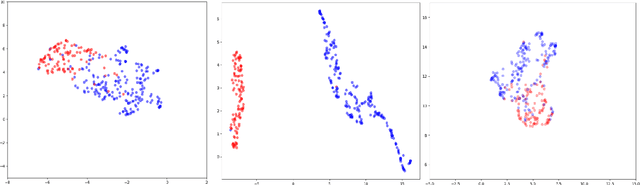
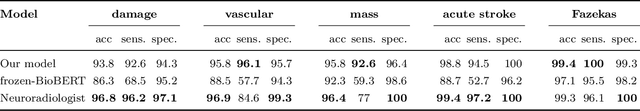
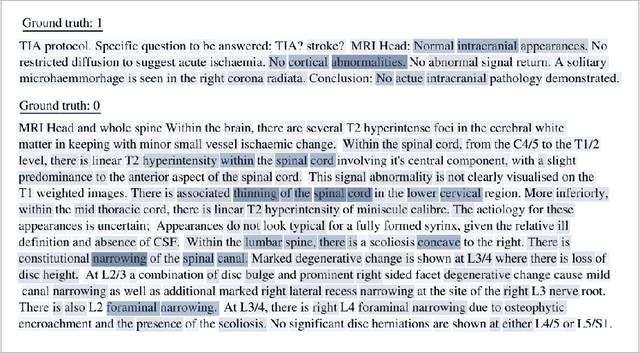
Abstract:Labelling large datasets for training high-capacity neural networks is a major obstacle to the development of deep learning-based medical imaging applications. Here we present a transformer-based network for magnetic resonance imaging (MRI) radiology report classification which automates this task by assigning image labels on the basis of free-text expert radiology reports. Our model's performance is comparable to that of an expert radiologist, and better than that of an expert physician, demonstrating the feasibility of this approach. We make code available online for researchers to label their own MRI datasets for medical imaging applications.
Analysis of an Automated Machine Learning Approach in Brain Predictive Modelling: A data-driven approach to Predict Brain Age from Cortical Anatomical Measures
Oct 08, 2019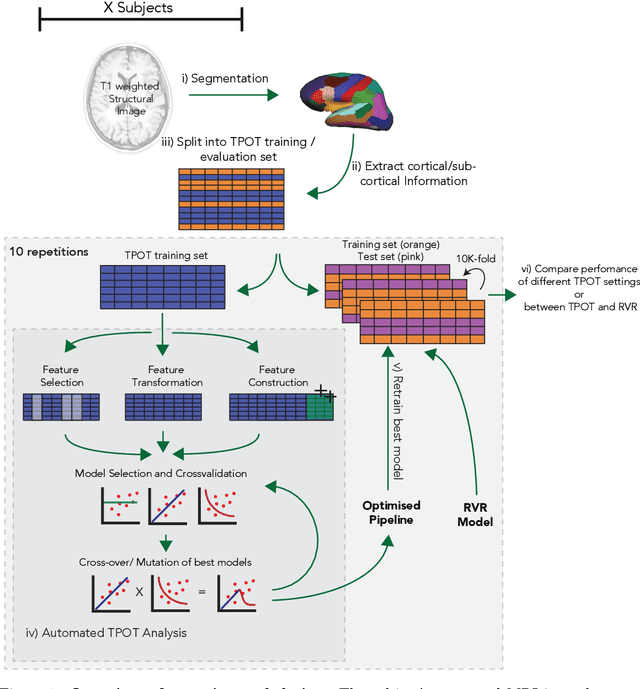


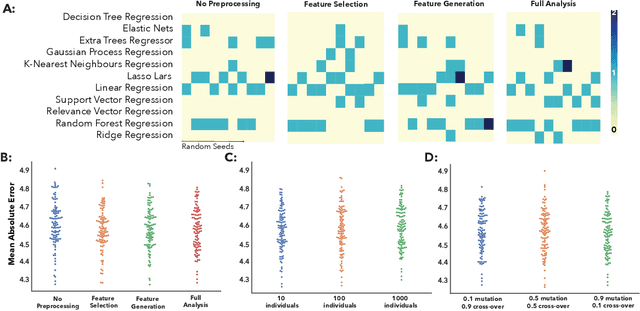
Abstract:The use of machine learning (ML) algorithms has significantly increased in neuroscience. However, from the vast extent of possible ML algorithms, which one is the optimal model to predict the target variable? What are the hyperparameters for such a model? Given the plethora of possible answers to these questions, in the last years, automated machine learning (autoML) has been gaining attention. Here, we apply an autoML library called TPOT which uses a tree-based representation of machine learning pipelines and conducts a genetic-programming based approach to find the model and its hyperparameters that more closely predicts the subject's true age. To explore autoML and evaluate its efficacy within neuroimaging datasets, we chose a problem that has been the focus of previous extensive study: brain age prediction. Without any prior knowledge, TPOT was able to scan through the model space and create pipelines that outperformed the state-of-the-art accuracy for Freesurfer-based models using only thickness and volume information for anatomical structure. In particular, we compared the performance of TPOT (mean accuracy error (MAE): $4.612 \pm .124$ years) and a Relevance Vector Regression (MAE $5.474 \pm .140$ years). TPOT also suggested interesting combinations of models that do not match the current most used models for brain prediction but generalise well to unseen data. AutoML showed promising results as a data-driven approach to find optimal models for neuroimaging applications.
Classification of Major Depressive Disorder via Multi-Site Weighted LASSO Model
Jun 03, 2017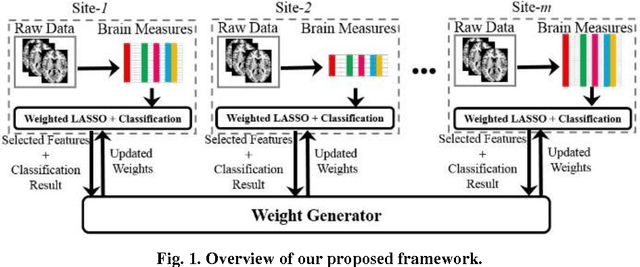
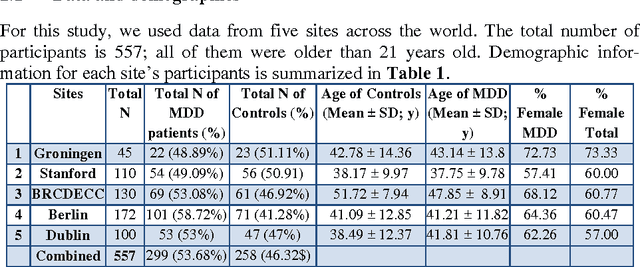
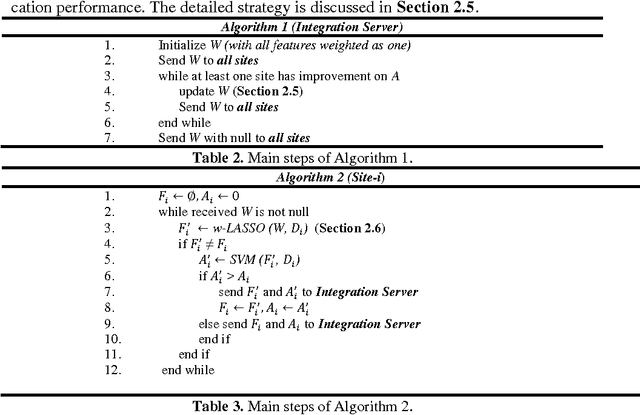
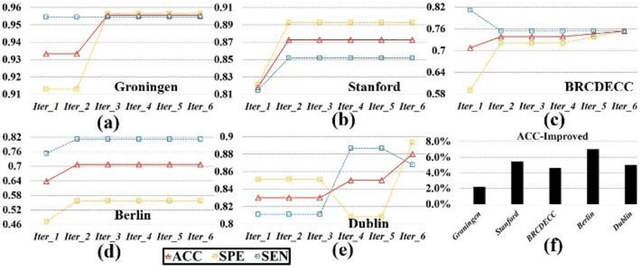
Abstract:Large-scale collaborative analysis of brain imaging data, in psychiatry and neu-rology, offers a new source of statistical power to discover features that boost ac-curacy in disease classification, differential diagnosis, and outcome prediction. However, due to data privacy regulations or limited accessibility to large datasets across the world, it is challenging to efficiently integrate distributed information. Here we propose a novel classification framework through multi-site weighted LASSO: each site performs an iterative weighted LASSO for feature selection separately. Within each iteration, the classification result and the selected features are collected to update the weighting parameters for each feature. This new weight is used to guide the LASSO process at the next iteration. Only the fea-tures that help to improve the classification accuracy are preserved. In tests on da-ta from five sites (299 patients with major depressive disorder (MDD) and 258 normal controls), our method boosted classification accuracy for MDD by 4.9% on average. This result shows the potential of the proposed new strategy as an ef-fective and practical collaborative platform for machine learning on large scale distributed imaging and biobank data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge