Robert Leech
GAMBAS: Generalised-Hilbert Mamba for Super-resolution of Paediatric Ultra-Low-Field MRI
Apr 06, 2025Abstract:Magnetic resonance imaging (MRI) is critical for neurodevelopmental research, however access to high-field (HF) systems in low- and middle-income countries is severely hindered by their cost. Ultra-low-field (ULF) systems mitigate such issues of access inequality, however their diminished signal-to-noise ratio limits their applicability for research and clinical use. Deep-learning approaches can enhance the quality of scans acquired at lower field strengths at no additional cost. For example, Convolutional neural networks (CNNs) fused with transformer modules have demonstrated a remarkable ability to capture both local information and long-range context. Unfortunately, the quadratic complexity of transformers leads to an undesirable trade-off between long-range sensitivity and local precision. We propose a hybrid CNN and state-space model (SSM) architecture featuring a novel 3D to 1D serialisation (GAMBAS), which learns long-range context without sacrificing spatial precision. We exhibit improved performance compared to other state-of-the-art medical image-to-image translation models.
SIM: Surface-based fMRI Analysis for Inter-Subject Multimodal Decoding from Movie-Watching Experiments
Jan 27, 2025Abstract:Current AI frameworks for brain decoding and encoding, typically train and test models within the same datasets. This limits their utility for brain computer interfaces (BCI) or neurofeedback, for which it would be useful to pool experiences across individuals to better simulate stimuli not sampled during training. A key obstacle to model generalisation is the degree of variability of inter-subject cortical organisation, which makes it difficult to align or compare cortical signals across participants. In this paper we address this through the use of surface vision transformers, which build a generalisable model of cortical functional dynamics, through encoding the topography of cortical networks and their interactions as a moving image across a surface. This is then combined with tri-modal self-supervised contrastive (CLIP) alignment of audio, video, and fMRI modalities to enable the retrieval of visual and auditory stimuli from patterns of cortical activity (and vice-versa). We validate our approach on 7T task-fMRI data from 174 healthy participants engaged in the movie-watching experiment from the Human Connectome Project (HCP). Results show that it is possible to detect which movie clips an individual is watching purely from their brain activity, even for individuals and movies not seen during training. Further analysis of attention maps reveals that our model captures individual patterns of brain activity that reflect semantic and visual systems. This opens the door to future personalised simulations of brain function. Code & pre-trained models will be made available at https://github.com/metrics-lab/sim, processed data for training will be available upon request at https://gin.g-node.org/Sdahan30/sim.
Transformer-based normative modelling for anomaly detection of early schizophrenia
Dec 08, 2022Abstract:Despite the impact of psychiatric disorders on clinical health, early-stage diagnosis remains a challenge. Machine learning studies have shown that classifiers tend to be overly narrow in the diagnosis prediction task. The overlap between conditions leads to high heterogeneity among participants that is not adequately captured by classification models. To address this issue, normative approaches have surged as an alternative method. By using a generative model to learn the distribution of healthy brain data patterns, we can identify the presence of pathologies as deviations or outliers from the distribution learned by the model. In particular, deep generative models showed great results as normative models to identify neurological lesions in the brain. However, unlike most neurological lesions, psychiatric disorders present subtle changes widespread in several brain regions, making these alterations challenging to identify. In this work, we evaluate the performance of transformer-based normative models to detect subtle brain changes expressed in adolescents and young adults. We trained our model on 3D MRI scans of neurotypical individuals (N=1,765). Then, we obtained the likelihood of neurotypical controls and psychiatric patients with early-stage schizophrenia from an independent dataset (N=93) from the Human Connectome Project. Using the predicted likelihood of the scans as a proxy for a normative score, we obtained an AUROC of 0.82 when assessing the difference between controls and individuals with early-stage schizophrenia. Our approach surpassed recent normative methods based on brain age and Gaussian Process, showing the promising use of deep generative models to help in individualised analyses.
Causal Autoregressive Flows
Nov 04, 2020



Abstract:Two apparently unrelated fields -- normalizing flows and causality -- have recently received considerable attention in the machine learning community. In this work, we highlight an intrinsic correspondence between a simple family of flows and identifiable causal models. We exploit the fact that autoregressive flow architectures define an ordering over variables, analogous to a causal ordering, to show that they are well-suited to performing a range of causal inference tasks. First, we show that causal models derived from both affine and additive flows are identifiable. This provides a generalization of the additive noise model well-known in causal discovery. Second, we derive a bivariate measure of causal direction based on likelihood ratios, leveraging the fact that flow models estimate normalized log-densities of data. Such likelihood ratios have well-known optimality properties in finite-sample inference. Third, we demonstrate that the invertibility of flows naturally allows for direct evaluation of both interventional and counterfactual queries. Finally, throughout a series of experiments on synthetic and real data, the proposed method is shown to outperform current approaches for causal discovery as well as making accurate interventional and counterfactual predictions.
Bayesian optimization for automatic design of face stimuli
Jul 20, 2020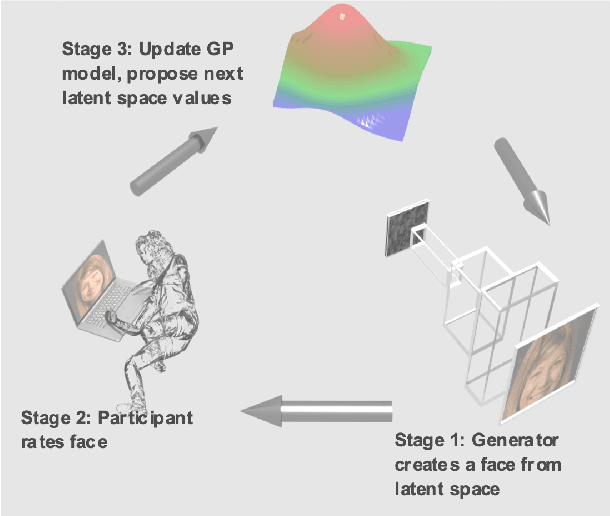
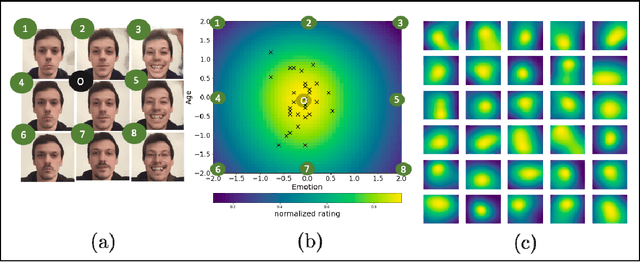
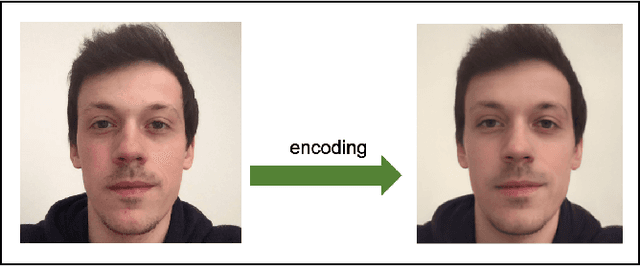

Abstract:Investigating the cognitive and neural mechanisms involved with face processing is a fundamental task in modern neuroscience and psychology. To date, the majority of such studies have focused on the use of pre-selected stimuli. The absence of personalized stimuli presents a serious limitation as it fails to account for how each individual face processing system is tuned to cultural embeddings or how it is disrupted in disease. In this work, we propose a novel framework which combines generative adversarial networks (GANs) with Bayesian optimization to identify individual response patterns to many different faces. Formally, we employ Bayesian optimization to efficiently search the latent space of state-of-the-art GAN models, with the aim to automatically generate novel faces, to maximize an individual subject's response. We present results from a web-based proof-of-principle study, where participants rated images of themselves generated via performing Bayesian optimization over the latent space of a GAN. We show how the algorithm can efficiently locate an individual's optimal face while mapping out their response across different semantic transformations of a face; inter-individual analyses suggest how the approach can provide rich information about individual differences in face processing.
Analysis of an Automated Machine Learning Approach in Brain Predictive Modelling: A data-driven approach to Predict Brain Age from Cortical Anatomical Measures
Oct 08, 2019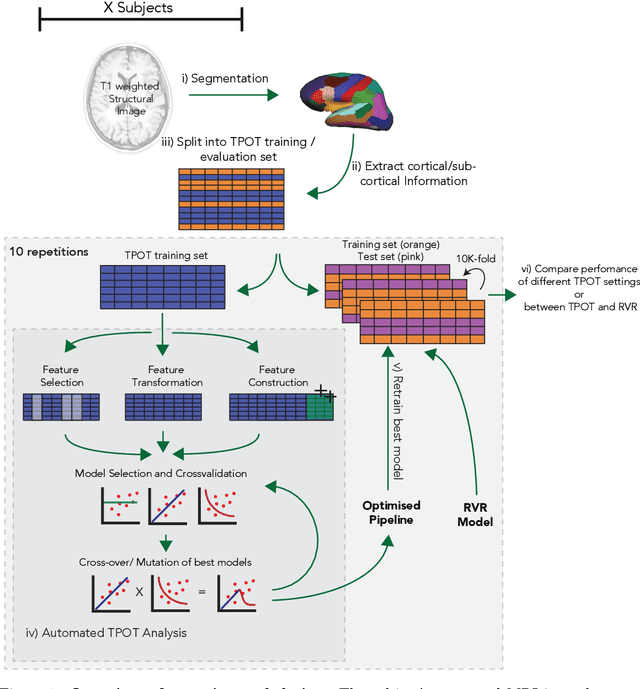


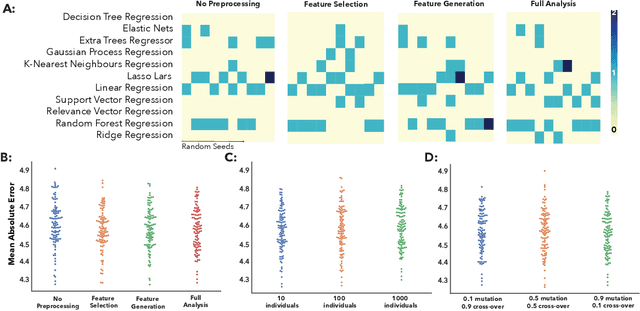
Abstract:The use of machine learning (ML) algorithms has significantly increased in neuroscience. However, from the vast extent of possible ML algorithms, which one is the optimal model to predict the target variable? What are the hyperparameters for such a model? Given the plethora of possible answers to these questions, in the last years, automated machine learning (autoML) has been gaining attention. Here, we apply an autoML library called TPOT which uses a tree-based representation of machine learning pipelines and conducts a genetic-programming based approach to find the model and its hyperparameters that more closely predicts the subject's true age. To explore autoML and evaluate its efficacy within neuroimaging datasets, we chose a problem that has been the focus of previous extensive study: brain age prediction. Without any prior knowledge, TPOT was able to scan through the model space and create pipelines that outperformed the state-of-the-art accuracy for Freesurfer-based models using only thickness and volume information for anatomical structure. In particular, we compared the performance of TPOT (mean accuracy error (MAE): $4.612 \pm .124$ years) and a Relevance Vector Regression (MAE $5.474 \pm .140$ years). TPOT also suggested interesting combinations of models that do not match the current most used models for brain prediction but generalise well to unseen data. AutoML showed promising results as a data-driven approach to find optimal models for neuroimaging applications.
Streaming regularization parameter selection via stochastic gradient descent
Nov 02, 2016Abstract:We propose a framework to perform streaming covariance selection. Our approach employs regularization constraints where a time-varying sparsity parameter is iteratively estimated via stochastic gradient descent. This allows for the regularization parameter to be efficiently learnt in an online manner. The proposed framework is developed for linear regression models and extended to graphical models via neighbourhood selection. Under mild assumptions, we are able to obtain convergence results in a non-stochastic setting. The capabilities of such an approach are demonstrated using both synthetic data as well as neuroimaging data.
Text-mining the NeuroSynth corpus using Deep Boltzmann Machines
May 01, 2016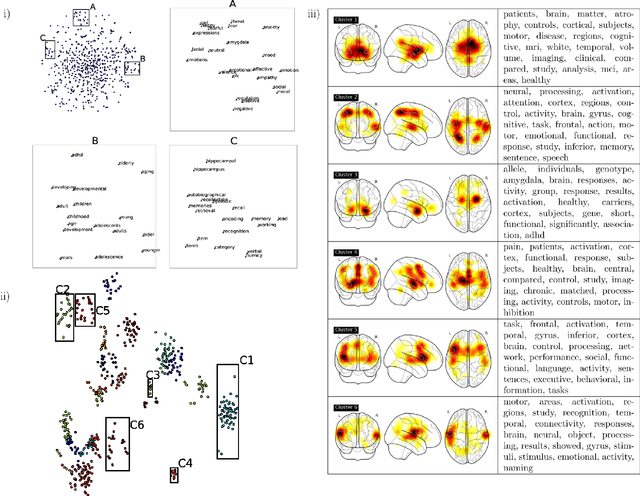
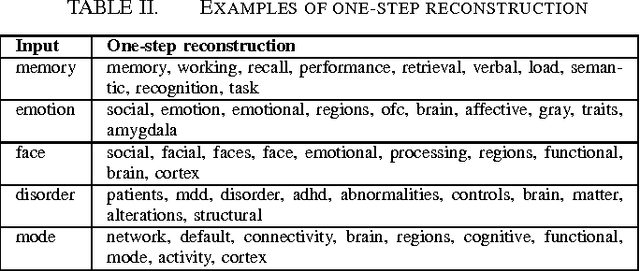
Abstract:Large-scale automated meta-analysis of neuroimaging data has recently established itself as an important tool in advancing our understanding of human brain function. This research has been pioneered by NeuroSynth, a database collecting both brain activation coordinates and associated text across a large cohort of neuroimaging research papers. One of the fundamental aspects of such meta-analysis is text-mining. To date, word counts and more sophisticated methods such as Latent Dirichlet Allocation have been proposed. In this work we present an unsupervised study of the NeuroSynth text corpus using Deep Boltzmann Machines (DBMs). The use of DBMs yields several advantages over the aforementioned methods, principal among which is the fact that it yields both word and document embeddings in a high-dimensional vector space. Such embeddings serve to facilitate the use of traditional machine learning techniques on the text corpus. The proposed DBM model is shown to learn embeddings with a clear semantic structure.
Stopping criteria for boosting automatic experimental design using real-time fMRI with Bayesian optimization
Mar 22, 2016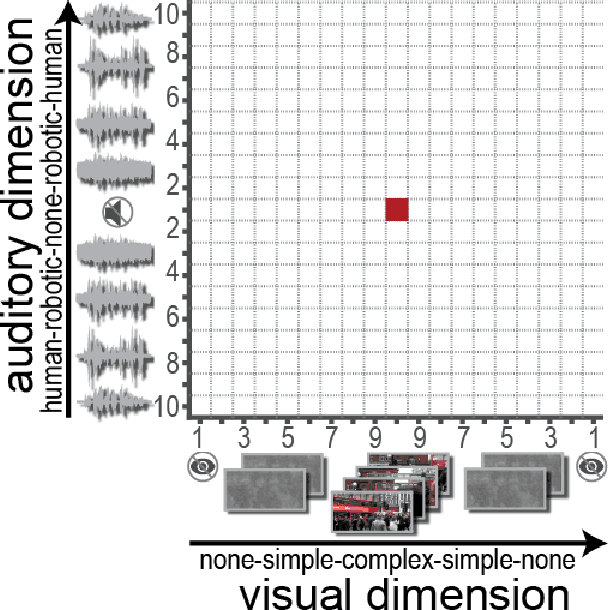
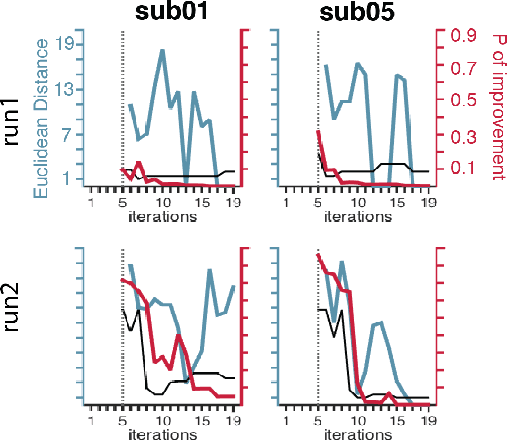
Abstract:Bayesian optimization has been proposed as a practical and efficient tool through which to tune parameters in many difficult settings. Recently, such techniques have been combined with real-time fMRI to propose a novel framework which turns on its head the conventional functional neuroimaging approach. This closed-loop method automatically designs the optimal experiment to evoke a desired target brain pattern. One of the challenges associated with extending such methods to real-time brain imaging is the need for adequate stopping criteria, an aspect of Bayesian optimization which has received limited attention. In light of high scanning costs and limited attentional capacities of subjects an accurate and reliable stopping criteria is essential. In order to address this issue we propose and empirically study the performance of two stopping criteria.
Measuring the functional connectome "on-the-fly": towards a new control signal for fMRI-based brain-computer interfaces
Feb 08, 2015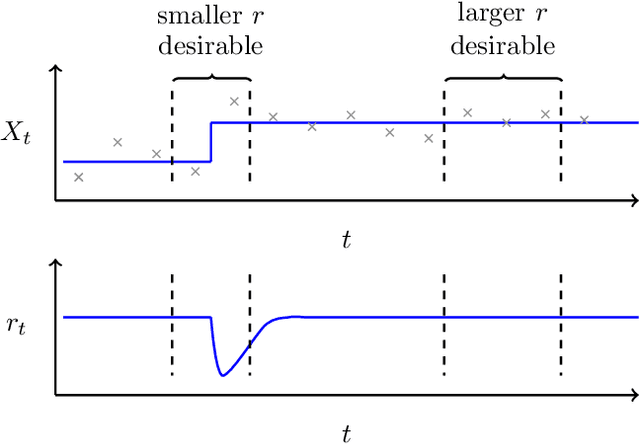
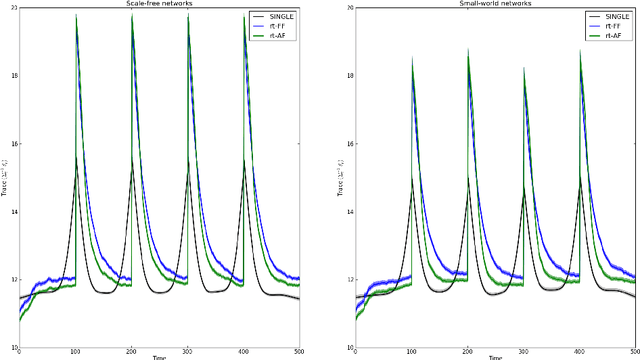
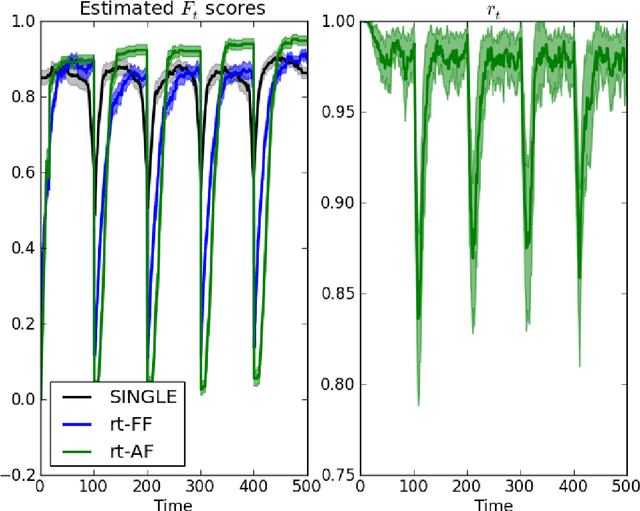
Abstract:There has been an explosion of interest in functional Magnetic Resonance Imaging (MRI) during the past two decades. Naturally, this has been accompanied by many major advances in the understanding of the human connectome. These advances have served to pose novel challenges as well as open new avenues for research. One of the most promising and exciting of such avenues is the study of functional MRI in real-time. Such studies have recently gained momentum and have been applied in a wide variety of settings; ranging from training of healthy subjects to self-regulate neuronal activity to being suggested as potential treatments for clinical populations. To date, the vast majority of these studies have focused on a single region at a time. This is due in part to the many challenges faced when estimating dynamic functional connectivity networks in real-time. In this work we propose a novel methodology with which to accurately track changes in functional connectivity networks in real-time. We adapt the recently proposed SINGLE algorithm for estimating sparse and temporally homo- geneous dynamic networks to be applicable in real-time. The proposed method is applied to motor task data from the Human Connectome Project as well as to real-time data ob- tained while exploring a virtual environment. We show that the algorithm is able to estimate significant task-related changes in network structure quickly enough to be useful in future brain-computer interface applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge