Romy Lorenz
Bayesian optimization for automatic design of face stimuli
Jul 20, 2020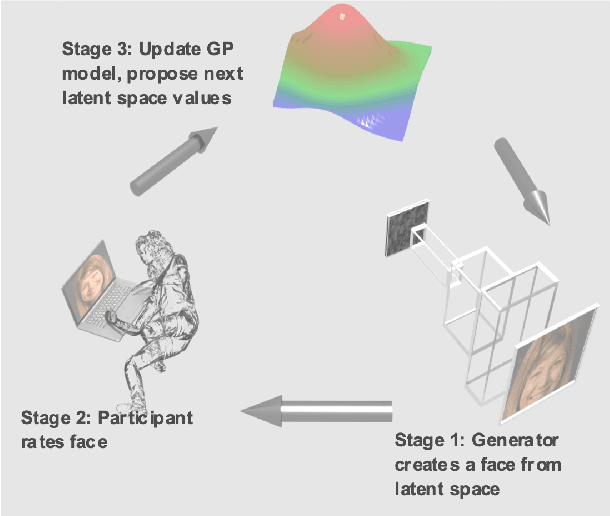
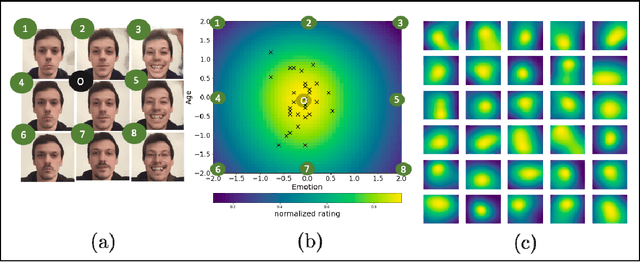
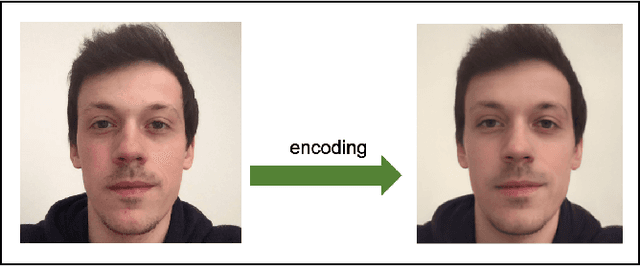

Abstract:Investigating the cognitive and neural mechanisms involved with face processing is a fundamental task in modern neuroscience and psychology. To date, the majority of such studies have focused on the use of pre-selected stimuli. The absence of personalized stimuli presents a serious limitation as it fails to account for how each individual face processing system is tuned to cultural embeddings or how it is disrupted in disease. In this work, we propose a novel framework which combines generative adversarial networks (GANs) with Bayesian optimization to identify individual response patterns to many different faces. Formally, we employ Bayesian optimization to efficiently search the latent space of state-of-the-art GAN models, with the aim to automatically generate novel faces, to maximize an individual subject's response. We present results from a web-based proof-of-principle study, where participants rated images of themselves generated via performing Bayesian optimization over the latent space of a GAN. We show how the algorithm can efficiently locate an individual's optimal face while mapping out their response across different semantic transformations of a face; inter-individual analyses suggest how the approach can provide rich information about individual differences in face processing.
Streaming regularization parameter selection via stochastic gradient descent
Nov 02, 2016Abstract:We propose a framework to perform streaming covariance selection. Our approach employs regularization constraints where a time-varying sparsity parameter is iteratively estimated via stochastic gradient descent. This allows for the regularization parameter to be efficiently learnt in an online manner. The proposed framework is developed for linear regression models and extended to graphical models via neighbourhood selection. Under mild assumptions, we are able to obtain convergence results in a non-stochastic setting. The capabilities of such an approach are demonstrated using both synthetic data as well as neuroimaging data.
Text-mining the NeuroSynth corpus using Deep Boltzmann Machines
May 01, 2016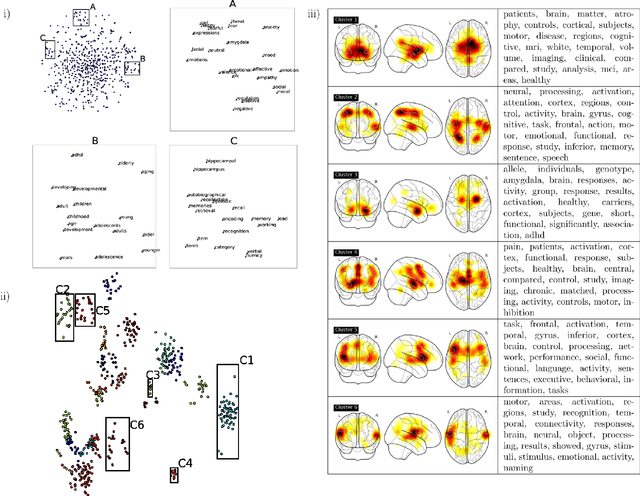
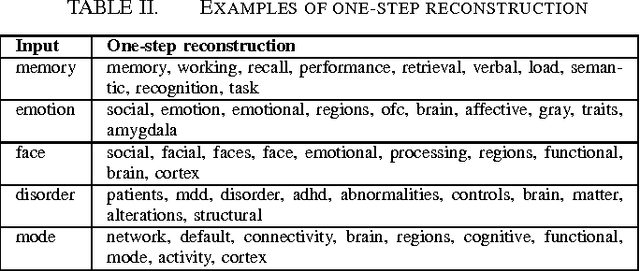
Abstract:Large-scale automated meta-analysis of neuroimaging data has recently established itself as an important tool in advancing our understanding of human brain function. This research has been pioneered by NeuroSynth, a database collecting both brain activation coordinates and associated text across a large cohort of neuroimaging research papers. One of the fundamental aspects of such meta-analysis is text-mining. To date, word counts and more sophisticated methods such as Latent Dirichlet Allocation have been proposed. In this work we present an unsupervised study of the NeuroSynth text corpus using Deep Boltzmann Machines (DBMs). The use of DBMs yields several advantages over the aforementioned methods, principal among which is the fact that it yields both word and document embeddings in a high-dimensional vector space. Such embeddings serve to facilitate the use of traditional machine learning techniques on the text corpus. The proposed DBM model is shown to learn embeddings with a clear semantic structure.
Stopping criteria for boosting automatic experimental design using real-time fMRI with Bayesian optimization
Mar 22, 2016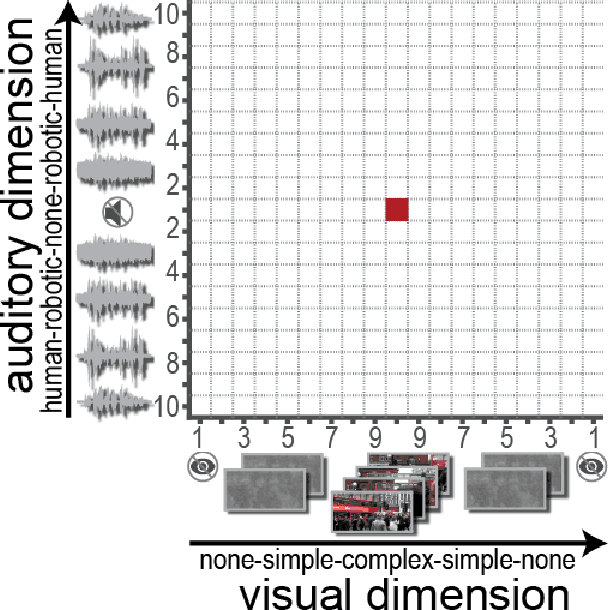
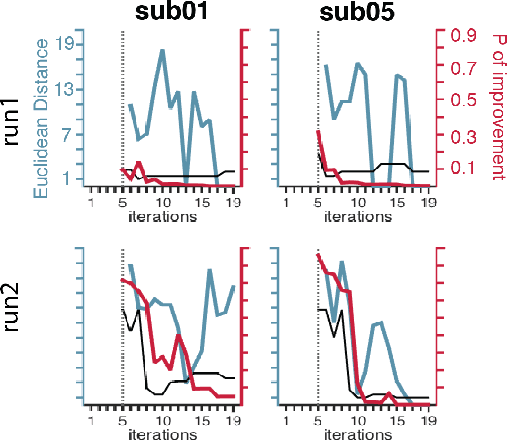
Abstract:Bayesian optimization has been proposed as a practical and efficient tool through which to tune parameters in many difficult settings. Recently, such techniques have been combined with real-time fMRI to propose a novel framework which turns on its head the conventional functional neuroimaging approach. This closed-loop method automatically designs the optimal experiment to evoke a desired target brain pattern. One of the challenges associated with extending such methods to real-time brain imaging is the need for adequate stopping criteria, an aspect of Bayesian optimization which has received limited attention. In light of high scanning costs and limited attentional capacities of subjects an accurate and reliable stopping criteria is essential. In order to address this issue we propose and empirically study the performance of two stopping criteria.
Measuring the functional connectome "on-the-fly": towards a new control signal for fMRI-based brain-computer interfaces
Feb 08, 2015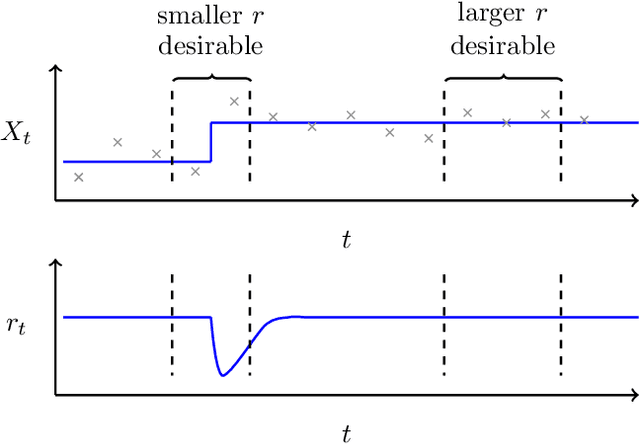
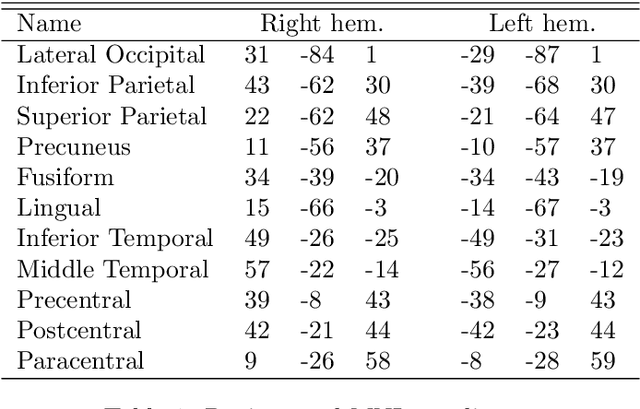
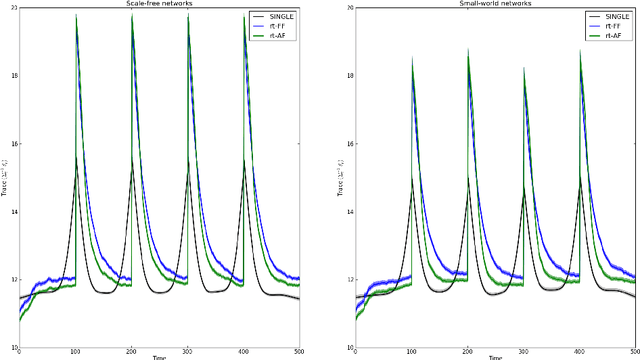
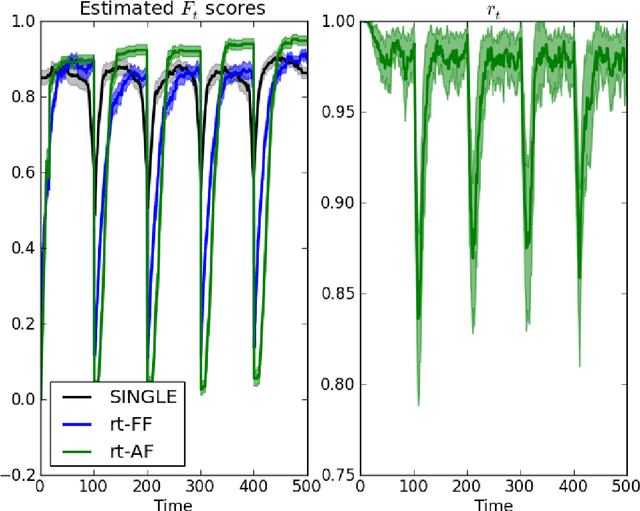
Abstract:There has been an explosion of interest in functional Magnetic Resonance Imaging (MRI) during the past two decades. Naturally, this has been accompanied by many major advances in the understanding of the human connectome. These advances have served to pose novel challenges as well as open new avenues for research. One of the most promising and exciting of such avenues is the study of functional MRI in real-time. Such studies have recently gained momentum and have been applied in a wide variety of settings; ranging from training of healthy subjects to self-regulate neuronal activity to being suggested as potential treatments for clinical populations. To date, the vast majority of these studies have focused on a single region at a time. This is due in part to the many challenges faced when estimating dynamic functional connectivity networks in real-time. In this work we propose a novel methodology with which to accurately track changes in functional connectivity networks in real-time. We adapt the recently proposed SINGLE algorithm for estimating sparse and temporally homo- geneous dynamic networks to be applicable in real-time. The proposed method is applied to motor task data from the Human Connectome Project as well as to real-time data ob- tained while exploring a virtual environment. We show that the algorithm is able to estimate significant task-related changes in network structure quickly enough to be useful in future brain-computer interface applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge