Jan Philipp Albrecht
Harnessing spatial homogeneity of neuroimaging data: patch individual filter layers for CNNs
Jul 23, 2020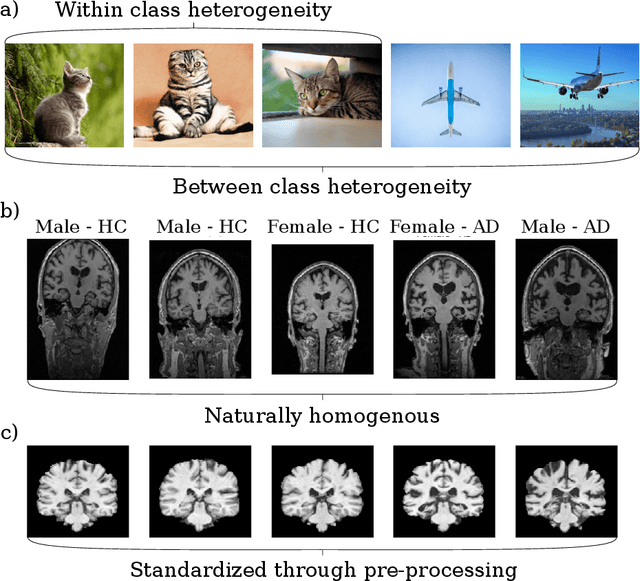
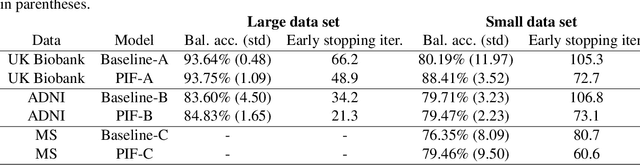
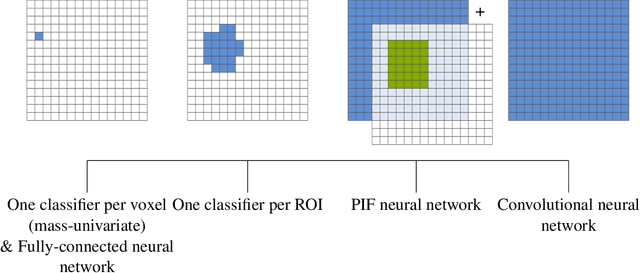
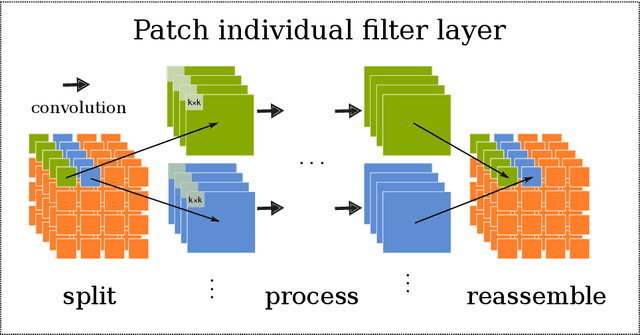
Abstract:Neuroimaging data, e.g. obtained from magnetic resonance imaging (MRI), is comparably homogeneous due to (1) the uniform structure of the brain and (2) additional efforts to spatially normalize the data to a standard template using linear and non-linear transformations. Convolutional neural networks (CNNs), in contrast, have been specifically designed for highly heterogeneous data, such as natural images, by sliding convolutional filters over different positions in an image. Here, we suggest a new CNN architecture that combines the idea of hierarchical abstraction in neural networks with a prior on the spatial homogeneity of neuroimaging data: Whereas early layers are trained globally using standard convolutional layers, we introduce for higher, more abstract layers patch individual filters (PIF). By learning filters in individual image regions (patches) without sharing weights, PIF layers can learn abstract features faster and with fewer samples. We thoroughly evaluated PIF layers for three different tasks and data sets, namely sex classification on UK Biobank data, Alzheimer's disease detection on ADNI data and multiple sclerosis detection on private hospital data. We demonstrate that CNNs using PIF layers result in higher accuracies, especially in low sample size settings, and need fewer training epochs for convergence. To the best of our knowledge, this is the first study which introduces a prior on brain MRI for CNN learning.
Harnessing spatial MRI normalization: patch individual filter layers for CNNs
Nov 14, 2019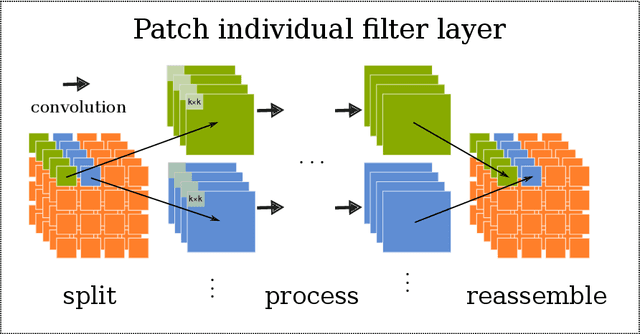
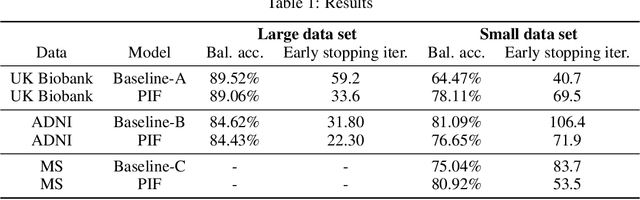
Abstract:Neuroimaging studies based on magnetic resonance imaging (MRI) typically employ rigorous forms of preprocessing. Images are spatially normalized to a standard template using linear and non-linear transformations. Thus, one can assume that a patch at location (x, y, height, width) contains the same brain region across the entire data set. Most analyses applied on brain MRI using convolutional neural networks (CNNs) ignore this distinction from natural images. Here, we suggest a new layer type called patch individual filter (PIF) layer, which trains higher-level filters locally as we assume that more abstract features are locally specific after spatial normalization. We evaluate PIF layers on three different tasks, namely sex classification as well as either Alzheimer's disease (AD) or multiple sclerosis (MS) detection. We demonstrate that CNNs using PIF layers outperform their counterparts in several, especially low sample size settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge