Hangfan Liu
Deep Learning Based Detection of Enlarged Perivascular Spaces on Brain MRI
Sep 27, 2022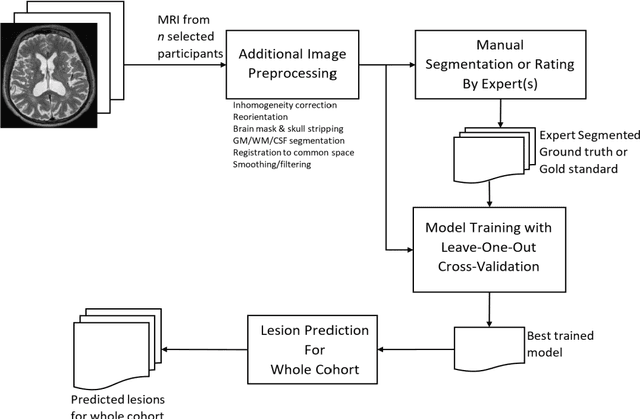
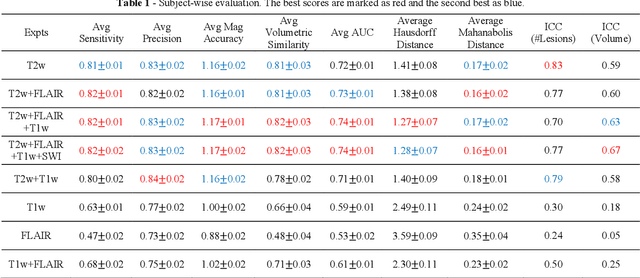
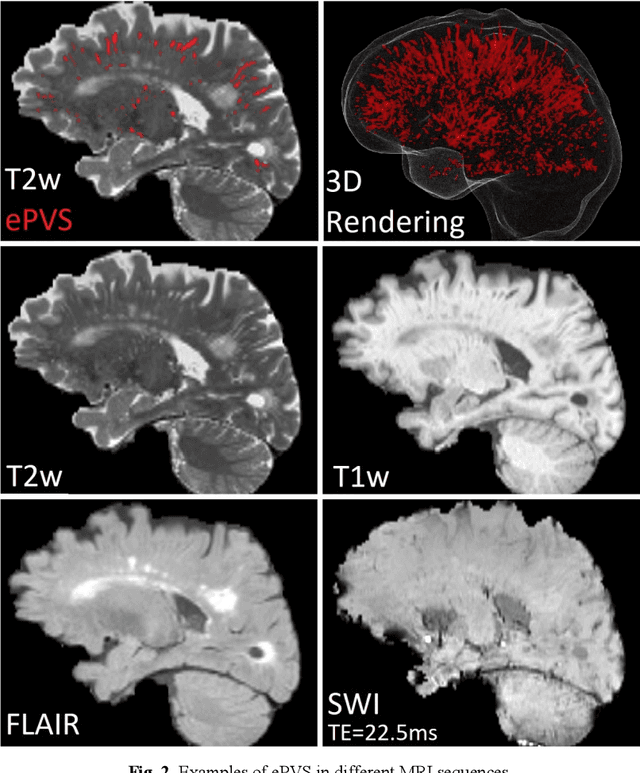
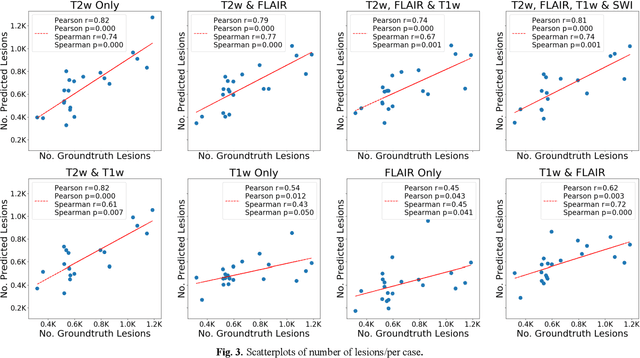
Abstract:Deep learning has been demonstrated effective in many neuroimaging applications. However, in many scenarios the number of imaging sequences capturing information related to small vessel disease lesions is insufficient to support data-driven techniques. Additionally, cohort-based studies may not always have the optimal or essential imaging sequences for accurate lesion detection. Therefore, it is necessary to determine which of these imaging sequences are essential for accurate detection. In this study we aimed to find the optimal combination of magnetic resonance imaging (MRI) sequences for deep learning-based detection of enlarged perivascular spaces (ePVS). To this end, we implemented an effective light-weight U-Net adapted for ePVS detection and comprehensively investigated different combinations of information from susceptibility weighted imaging (SWI), fluid-attenuated inversion recovery (FLAIR), T1-weighted (T1w) and T2-weighted (T2w) MRI sequences. We conclude that T2w MRI is the most important for accurate ePVS detection, and the incorporation of SWI, FLAIR and T1w MRI in the deep neural network could make insignificant improvements in accuracy.
COLA-Net: Collaborative Attention Network for Image Restoration
Mar 10, 2021



Abstract:Local and non-local attention-based methods have been well studied in various image restoration tasks while leading to promising performance. However, most of the existing methods solely focus on one type of attention mechanism (local or non-local). Furthermore, by exploiting the self-similarity of natural images, existing pixel-wise non-local attention operations tend to give rise to deviations in the process of characterizing long-range dependence due to image degeneration. To overcome these problems, in this paper we propose a novel collaborative attention network (COLA-Net) for image restoration, as the first attempt to combine local and non-local attention mechanisms to restore image content in the areas with complex textures and with highly repetitive details respectively. In addition, an effective and robust patch-wise non-local attention model is developed to capture long-range feature correspondences through 3D patches. Extensive experiments on synthetic image denoising, real image denoising and compression artifact reduction tasks demonstrate that our proposed COLA-Net is able to achieve state-of-the-art performance in both peak signal-to-noise ratio and visual perception, while maintaining an attractive computational complexity. The source code is available on https://github.com/MC-E/COLA-Net.
Feature-Fused Context-Encoding Network for Neuroanatomy Segmentation
May 07, 2019



Abstract:Automatic segmentation of fine-grained brain structures remains a challenging task. Current segmentation methods mainly utilize 2D and 3D deep neural networks. The 2D networks take image slices as input to produce coarse segmentation in less processing time, whereas the 3D networks take the whole image volumes to generated fine-detailed segmentation with more computational burden. In order to obtain accurate fine-grained segmentation efficiently, in this paper, we propose an end-to-end Feature-Fused Context-Encoding Network for brain structure segmentation from MR (magnetic resonance) images. Our model is implemented based on a 2D convolutional backbone, which integrates a 2D encoding module to acquire planar image features and a spatial encoding module to extract spatial context information. A global context encoding module is further introduced to capture global context semantics from the fused 2D encoding and spatial features. The proposed network aims to fully leverage the global anatomical prior knowledge learned from context semantics, which is represented by a structure-aware attention factor to recalibrate the outputs of the network. In this way, the network is guaranteed to be aware of the class-dependent feature maps to facilitate the segmentation. We evaluate our model on 2012 Brain Multi-Atlas Labelling Challenge dataset for 134 fine-grained structure segmentation. Besides, we validate our network on 27 coarse structure segmentation tasks. Experimental results have demonstrated that our model can achieve improved performance compared with the state-of-the-art approaches.
DeepSEED: 3D Squeeze-and-Excitation Encoder-Decoder ConvNets for Pulmonary Nodule Detection
Apr 06, 2019


Abstract:Pulmonary nodule detection plays an important role in lung cancer screening with low-dose computed tomography (CT) scans. Although promising performance has been achieved by deep learning based nodule detection methods, it remains challenging to build nodule detection networks with good generalization performance due to unbalanced positive and negative samples. In order to overcome this problem and further improve state-of-the-art region proposal network methods, we develop a novel deep 3D convolutional neural network with an Encoder-Decoder structure for pulmonary nodule detection. Particularly, we utilize a dynamically scaled cross entropy loss to reduce the false positive rate and compensate the significant data imbalance problem. We adopt the squeeze-and-excitation structure to learn effective image features and fully utilize channel inter-dependency. We have validated our method based on publicly available CT scans from LIDC/IDRI dataset and its subset LUNA16 with thinner slices. Ablation studies and experimental results have demonstrated that our method could outperform state-of-the-art nodule detection methods by a large margin, with an average FROC score of 86.2% on LUNA16, and an average FROC score of 77.3% on LIDC/IDRI dataset when trained on LUNA16 only.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge