Yuemeng Li
Versatile Medical Image Segmentation Learned from Multi-Source Datasets via Model Self-Disambiguation
Nov 17, 2023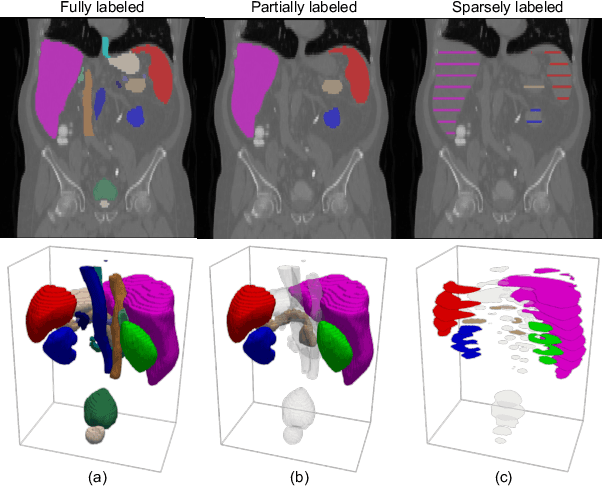
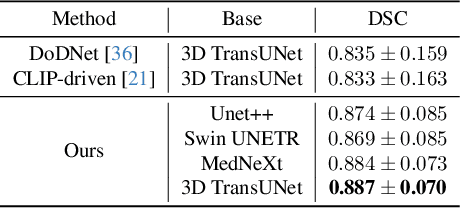
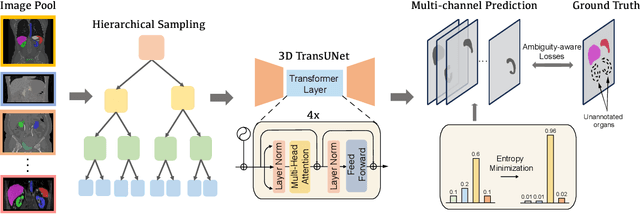

Abstract:A versatile medical image segmentation model applicable to imaging data collected with diverse equipment and protocols can facilitate model deployment and maintenance. However, building such a model typically requires a large, diverse, and fully annotated dataset, which is rarely available due to the labor-intensive and costly data curation. In this study, we develop a cost-efficient method by harnessing readily available data with partially or even sparsely annotated segmentation labels. We devise strategies for model self-disambiguation, prior knowledge incorporation, and imbalance mitigation to address challenges associated with inconsistently labeled data from various sources, including label ambiguity and imbalances across modalities, datasets, and segmentation labels. Experimental results on a multi-modal dataset compiled from eight different sources for abdominal organ segmentation have demonstrated our method's effectiveness and superior performance over alternative state-of-the-art methods, highlighting its potential for optimizing the use of existing annotated data and reducing the annotation efforts for new data to further enhance model capability.
Medical Image Segmentation with Domain Adaptation: A Survey
Nov 03, 2023



Abstract:Deep learning (DL) has shown remarkable success in various medical imaging data analysis applications. However, it remains challenging for DL models to achieve good generalization, especially when the training and testing datasets are collected at sites with different scanners, due to domain shift caused by differences in data distributions. Domain adaptation has emerged as an effective means to address this challenge by mitigating domain gaps in medical imaging applications. In this review, we specifically focus on domain adaptation approaches for DL-based medical image segmentation. We first present the motivation and background knowledge underlying domain adaptations, then provide a comprehensive review of domain adaptation applications in medical image segmentations, and finally discuss the challenges, limitations, and future research trends in the field to promote the methodology development of domain adaptation in the context of medical image segmentation. Our goal was to provide researchers with up-to-date references on the applications of domain adaptation in medical image segmentation studies.
Learning Apparent Diffusion Coefficient Maps from Undersampled Radial k-Space Diffusion-Weighted MRI in Mice using a Deep CNN-Transformer Model in Conjunction with a Monoexponential Model
Jul 06, 2022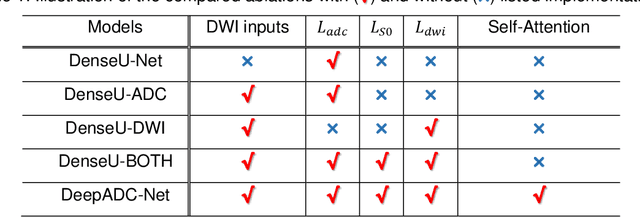
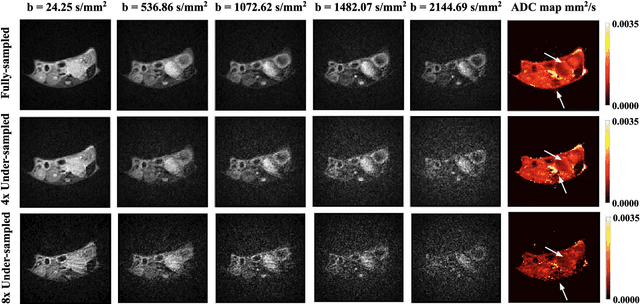
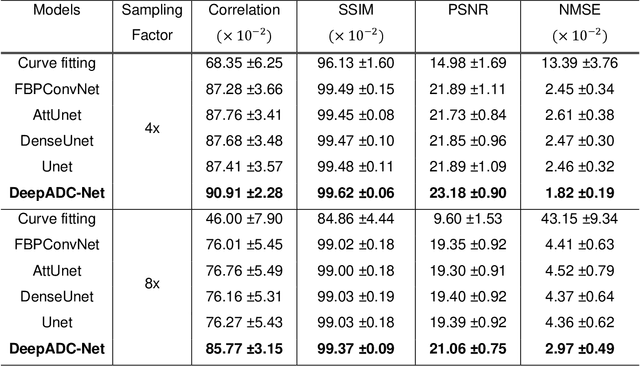
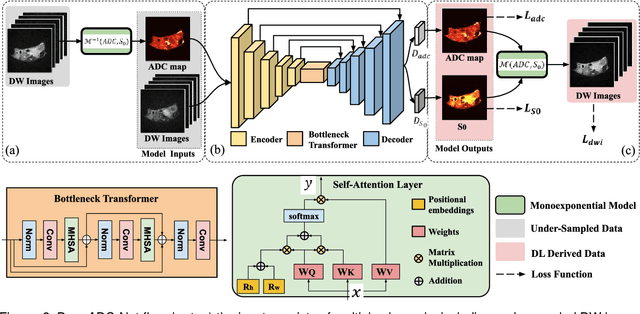
Abstract:Purpose: To accelerate radially sampled diffusion weighted spin-echo (Rad-DW-SE) acquisition method for generating high quality of apparent diffusion coefficient (ADC) maps. Methods: A deep learning method was developed to generate accurate ADC map reconstruction from undersampled DWI data acquired with the Rad-DW-SE method. The deep learning method integrates convolutional neural networks (CNNs) with vison transformers to generate high quality ADC maps from undersampled DWI data, regularized by a monoexponential ADC model fitting term. A model was trained on DWI data of 147 mice and evaluated on DWI data of 36 mice, with undersampling rates of 4x and 8x. Results: Ablation studies and experimental results have demonstrated that the proposed deep learning model can generate high quality ADC maps from undersampled DWI data, better than alternative deep learning methods under comparison, with their performance quantified on different levels of images, tumors, kidneys, and muscles. Conclusions: The deep learning method with integrated CNNs and transformers provides an effective means to accurately compute ADC maps from undersampled DWI data acquired with the Rad-DW-SE method.
Adaptive convolutional neural networks for k-space data interpolation in fast magnetic resonance imaging
Jun 09, 2020
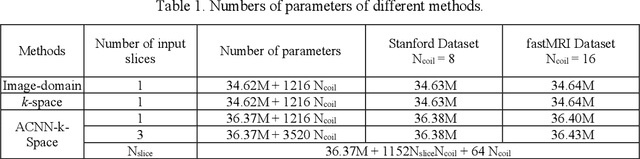

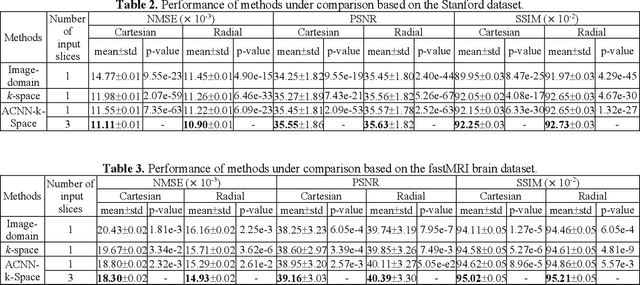
Abstract:Deep learning in k-space has demonstrated great potential for image reconstruction from undersampled k-space data in fast magnetic resonance imaging (MRI). However, existing deep learning-based image reconstruction methods typically apply weight-sharing convolutional neural networks (CNNs) to k-space data without taking into consideration the k-space data's spatial frequency properties, leading to ineffective learning of the image reconstruction models. Moreover, complementary information of spatially adjacent slices is often ignored in existing deep learning methods. To overcome such limitations, we develop a deep learning algorithm, referred to as adaptive convolutional neural networks for k-space data interpolation (ACNN-k-Space), which adopts a residual Encoder-Decoder network architecture to interpolate the undersampled k-space data by integrating spatially contiguous slices as multi-channel input, along with k-space data from multiple coils if available. The network is enhanced by self-attention layers to adaptively focus on k-space data at different spatial frequencies and channels. We have evaluated our method on two public datasets and compared it with state-of-the-art existing methods. Ablation studies and experimental results demonstrate that our method effectively reconstructs images from undersampled k-space data and achieves significantly better image reconstruction performance than current state-of-the-art techniques.
ACEnet: Anatomical Context-Encoding Network for Neuroanatomy Segmentation
Feb 13, 2020



Abstract:Segmentation of brain structures from magnetic resonance (MR) scans plays an important role in the quantification of brain morphology. Since 3D deep learning models suffer from high computational cost, 2D deep learning methods are favored for their computational efficiency. However, existing 2D deep learning methods are not equipped to effectively capture 3D spatial contextual information that is needed to achieve accurate brain structure segmentation. In order to overcome this limitation, we develop an Anatomical Context-Encoding Network (ACEnet) to incorporate 3D spatial and anatomical contexts in 2D convolutional neural networks (CNNs) for efficient and accurate segmentation of brain structures from MR scans, consisting of 1) an anatomical context encoding module to incorporate anatomical information in 2D CNNs, 2) a spatial context encoding module to integrate 3D image information in 2D CNNs, and 3) a skull stripping module to guide 2D CNNs to attend to the brain. Extensive experiments on three benchmark datasets have demonstrated that our method outperforms state-of-the-art alternative methods for brain structure segmentation in terms of both computational efficiency and segmentation accuracy.
Context-endcoding for neural network based skull stripping in magnetic resonance imaging
Oct 23, 2019


Abstract:Skull stripping is usually the first step for most brain analysisprocess in magnetic resonance images. A lot of deep learn-ing neural network based methods have been developed toachieve higher accuracy. Since the 3D deep learning modelssuffer from high computational cost and are subject to GPUmemory limit challenge, a variety of 2D deep learning meth-ods have been developed. However, existing 2D deep learn-ing methods are not equipped to effectively capture 3D se-mantic information that is needed to achieve higher accuracy.In this paper, we propose a context-encoding method to em-power the 2D network to capture the 3D context information.For the context-encoding method, firstly we encode the 2Dfeatures of original 2D network, secondly we encode the sub-volume of 3D MRI images, finally we fuse the encoded 2Dfeatures and 3D features with semantic encoding classifica-tion loss. To get computational efficiency, although we en-code the sub-volume of 3D MRI images instead of buildinga 3D neural network, extensive experiments on three bench-mark Datasets demonstrate our method can achieve superioraccuracy to state-of-the-art alternative methods with the dicescore 99.6% on NFBS and 99.09 % on LPBA40 and 99.17 %on OASIS.
Feature-Fused Context-Encoding Network for Neuroanatomy Segmentation
May 07, 2019



Abstract:Automatic segmentation of fine-grained brain structures remains a challenging task. Current segmentation methods mainly utilize 2D and 3D deep neural networks. The 2D networks take image slices as input to produce coarse segmentation in less processing time, whereas the 3D networks take the whole image volumes to generated fine-detailed segmentation with more computational burden. In order to obtain accurate fine-grained segmentation efficiently, in this paper, we propose an end-to-end Feature-Fused Context-Encoding Network for brain structure segmentation from MR (magnetic resonance) images. Our model is implemented based on a 2D convolutional backbone, which integrates a 2D encoding module to acquire planar image features and a spatial encoding module to extract spatial context information. A global context encoding module is further introduced to capture global context semantics from the fused 2D encoding and spatial features. The proposed network aims to fully leverage the global anatomical prior knowledge learned from context semantics, which is represented by a structure-aware attention factor to recalibrate the outputs of the network. In this way, the network is guaranteed to be aware of the class-dependent feature maps to facilitate the segmentation. We evaluate our model on 2012 Brain Multi-Atlas Labelling Challenge dataset for 134 fine-grained structure segmentation. Besides, we validate our network on 27 coarse structure segmentation tasks. Experimental results have demonstrated that our model can achieve improved performance compared with the state-of-the-art approaches.
DeepSEED: 3D Squeeze-and-Excitation Encoder-Decoder ConvNets for Pulmonary Nodule Detection
Apr 06, 2019


Abstract:Pulmonary nodule detection plays an important role in lung cancer screening with low-dose computed tomography (CT) scans. Although promising performance has been achieved by deep learning based nodule detection methods, it remains challenging to build nodule detection networks with good generalization performance due to unbalanced positive and negative samples. In order to overcome this problem and further improve state-of-the-art region proposal network methods, we develop a novel deep 3D convolutional neural network with an Encoder-Decoder structure for pulmonary nodule detection. Particularly, we utilize a dynamically scaled cross entropy loss to reduce the false positive rate and compensate the significant data imbalance problem. We adopt the squeeze-and-excitation structure to learn effective image features and fully utilize channel inter-dependency. We have validated our method based on publicly available CT scans from LIDC/IDRI dataset and its subset LUNA16 with thinner slices. Ablation studies and experimental results have demonstrated that our method could outperform state-of-the-art nodule detection methods by a large margin, with an average FROC score of 86.2% on LUNA16, and an average FROC score of 77.3% on LIDC/IDRI dataset when trained on LUNA16 only.
A Weakly Supervised Adaptive DenseNet for Classifying Thoracic Diseases and Identifying Abnormalities
Nov 05, 2018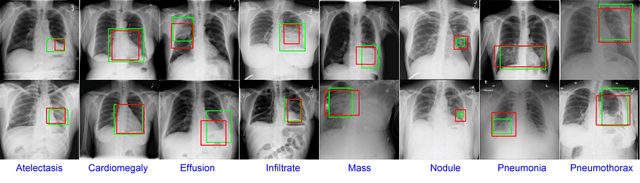
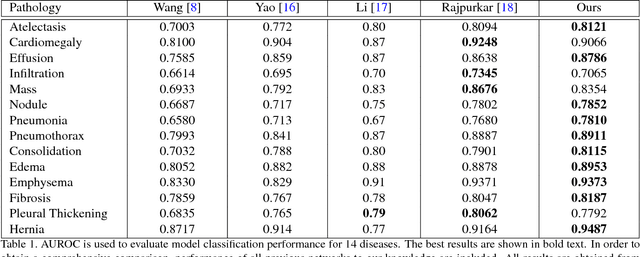
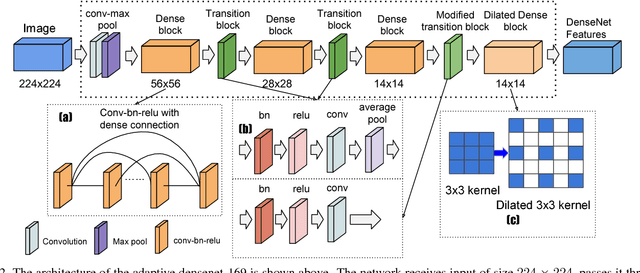
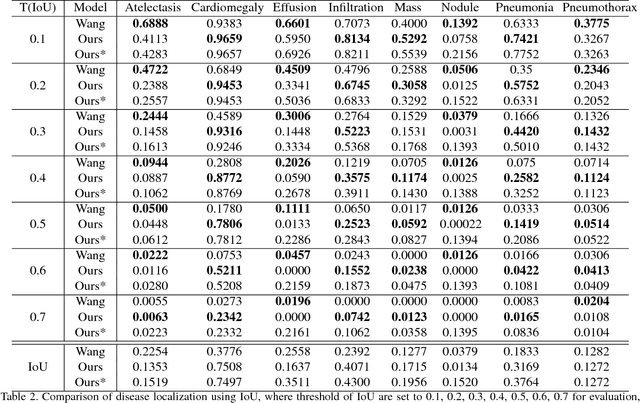
Abstract:We present a weakly supervised deep learning model for classifying thoracic diseases and identifying abnormalities in chest radiography. In this work, instead of learning from medical imaging data with region-level annotations, our model was merely trained on imaging data with image-level labels to classify diseases, and is able to identify abnormal image regions simultaneously. Our model consists of a customized pooling structure and an adaptive DenseNet front-end, which can effectively recognize possible disease features for classification and localization tasks. Our method has been validated on the publicly available ChestX-ray14 dataset. Experimental results have demonstrated that our classification and localization prediction performance achieved significant improvement over the previous models on the ChestX-ray14 dataset. In summary, our network can produce accurate disease classification and localization, which can potentially support clinical decisions.
On Spectral Analysis of Directed Signed Graphs
Dec 23, 2016



Abstract:It has been shown that the adjacency eigenspace of a network contains key information of its underlying structure. However, there has been no study on spectral analysis of the adjacency matrices of directed signed graphs. In this paper, we derive theoretical approximations of spectral projections from such directed signed networks using matrix perturbation theory. We use the derived theoretical results to study the influences of negative intra cluster and inter cluster directed edges on node spectral projections. We then develop a spectral clustering based graph partition algorithm, SC-DSG, and conduct evaluations on both synthetic and real datasets. Both theoretical analysis and empirical evaluation demonstrate the effectiveness of the proposed algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge