Hongming Li
ELEMENT: Episodic and Lifelong Exploration via Maximum Entropy
Dec 05, 2024Abstract:This paper proposes \emph{Episodic and Lifelong Exploration via Maximum ENTropy} (ELEMENT), a novel, multiscale, intrinsically motivated reinforcement learning (RL) framework that is able to explore environments without using any extrinsic reward and transfer effectively the learned skills to downstream tasks. We advance the state of the art in three ways. First, we propose a multiscale entropy optimization to take care of the fact that previous maximum state entropy, for lifelong exploration with millions of state observations, suffers from vanishing rewards and becomes very expensive computationally across iterations. Therefore, we add an episodic maximum entropy over each episode to speedup the search further. Second, we propose a novel intrinsic reward for episodic entropy maximization named \emph{average episodic state entropy} which provides the optimal solution for a theoretical upper bound of the episodic state entropy objective. Third, to speed the lifelong entropy maximization, we propose a $k$ nearest neighbors ($k$NN) graph to organize the estimation of the entropy and updating processes that reduces the computation substantially. Our ELEMENT significantly outperforms state-of-the-art intrinsic rewards in both episodic and lifelong setups. Moreover, it can be exploited in task-agnostic pre-training, collecting data for offline reinforcement learning, etc.
Revisiting a Pain in the Neck: Semantic Phrase Processing Benchmark for Language Models
May 05, 2024Abstract:We introduce LexBench, a comprehensive evaluation suite enabled to test language models (LMs) on ten semantic phrase processing tasks. Unlike prior studies, it is the first work to propose a framework from the comparative perspective to model the general semantic phrase (i.e., lexical collocation) and three fine-grained semantic phrases, including idiomatic expression, noun compound, and verbal construction. Thanks to \ourbenchmark, we assess the performance of 15 LMs across model architectures and parameter scales in classification, extraction, and interpretation tasks. Through the experiments, we first validate the scaling law and find that, as expected, large models excel better than the smaller ones in most tasks. Second, we investigate further through the scaling semantic relation categorization and find that few-shot LMs still lag behind vanilla fine-tuned models in the task. Third, through human evaluation, we find that the performance of strong models is comparable to the human level regarding semantic phrase processing. Our benchmarking findings can serve future research aiming to improve the generic capability of LMs on semantic phrase comprehension. Our source code and data are available at https://github.com/jacklanda/LexBench
Versatile Medical Image Segmentation Learned from Multi-Source Datasets via Model Self-Disambiguation
Nov 17, 2023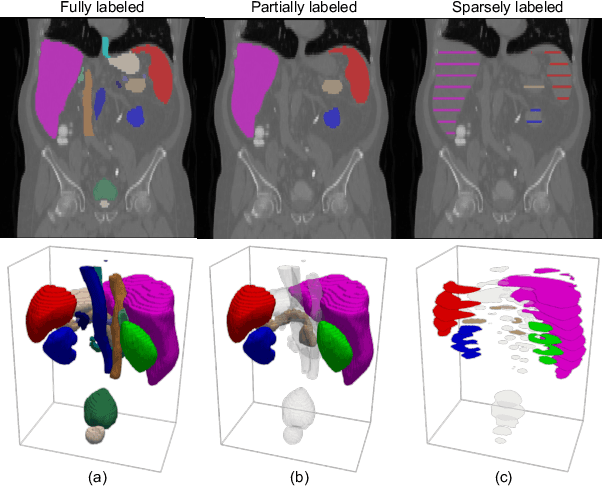
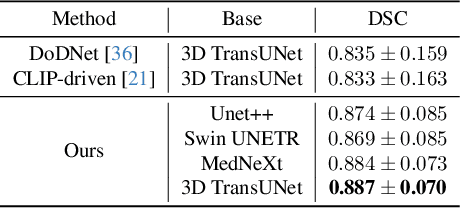
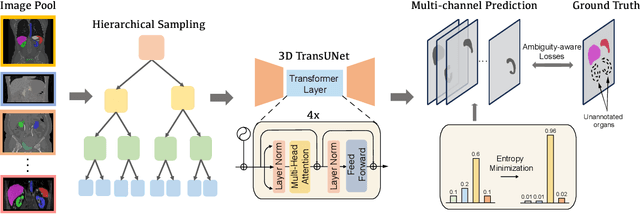

Abstract:A versatile medical image segmentation model applicable to imaging data collected with diverse equipment and protocols can facilitate model deployment and maintenance. However, building such a model typically requires a large, diverse, and fully annotated dataset, which is rarely available due to the labor-intensive and costly data curation. In this study, we develop a cost-efficient method by harnessing readily available data with partially or even sparsely annotated segmentation labels. We devise strategies for model self-disambiguation, prior knowledge incorporation, and imbalance mitigation to address challenges associated with inconsistently labeled data from various sources, including label ambiguity and imbalances across modalities, datasets, and segmentation labels. Experimental results on a multi-modal dataset compiled from eight different sources for abdominal organ segmentation have demonstrated our method's effectiveness and superior performance over alternative state-of-the-art methods, highlighting its potential for optimizing the use of existing annotated data and reducing the annotation efforts for new data to further enhance model capability.
SurfNN: Joint Reconstruction of Multiple Cortical Surfaces from Magnetic Resonance Images
Mar 06, 2023Abstract:To achieve fast, robust, and accurate reconstruction of the human cortical surfaces from 3D magnetic resonance images (MRIs), we develop a novel deep learning-based framework, referred to as SurfNN, to reconstruct simultaneously both inner (between white matter and gray matter) and outer (pial) surfaces from MRIs. Different from existing deep learning-based cortical surface reconstruction methods that either reconstruct the cortical surfaces separately or neglect the interdependence between the inner and outer surfaces, SurfNN reconstructs both the inner and outer cortical surfaces jointly by training a single network to predict a midthickness surface that lies at the center of the inner and outer cortical surfaces. The input of SurfNN consists of a 3D MRI and an initialization of the midthickness surface that is represented both implicitly as a 3D distance map and explicitly as a triangular mesh with spherical topology, and its output includes both the inner and outer cortical surfaces, as well as the midthickness surface. The method has been evaluated on a large-scale MRI dataset and demonstrated competitive cortical surface reconstruction performance.
Deep Clustering Survival Machines with Interpretable Expert Distributions
Jan 27, 2023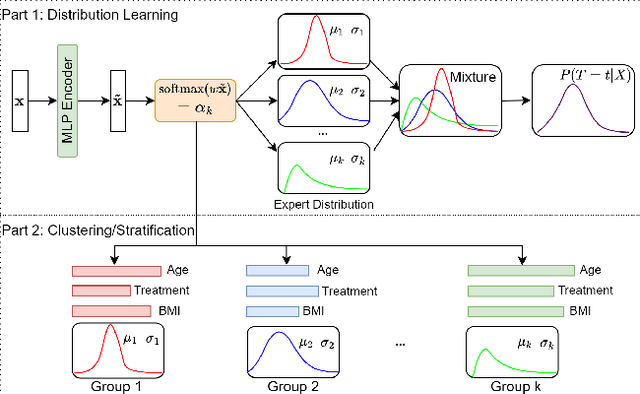
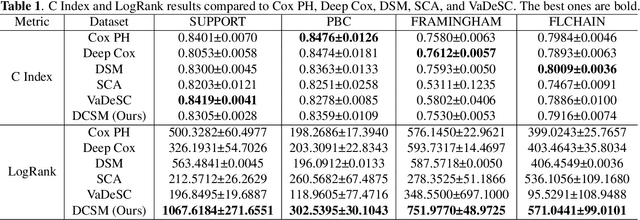
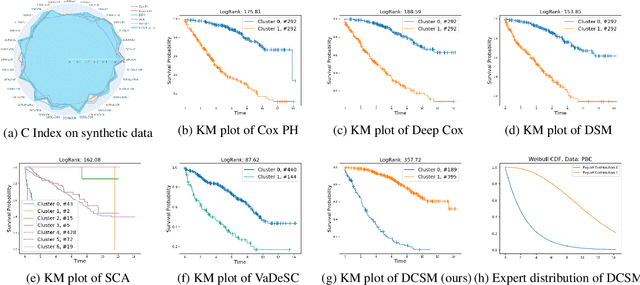
Abstract:Conventional survival analysis methods are typically ineffective to characterize heterogeneity in the population while such information can be used to assist predictive modeling. In this study, we propose a hybrid survival analysis method, referred to as deep clustering survival machines, that combines the discriminative and generative mechanisms. Similar to the mixture models, we assume that the timing information of survival data is generatively described by a mixture of certain numbers of parametric distributions, i.e., expert distributions. We learn weights of the expert distributions for individual instances according to their features discriminatively such that each instance's survival information can be characterized by a weighted combination of the learned constant expert distributions. This method also facilitates interpretable subgrouping/clustering of all instances according to their associated expert distributions. Extensive experiments on both real and synthetic datasets have demonstrated that the method is capable of obtaining promising clustering results and competitive time-to-event predicting performance.
The Conditional Cauchy-Schwarz Divergence with Applications to Time-Series Data and Sequential Decision Making
Jan 21, 2023Abstract:The Cauchy-Schwarz (CS) divergence was developed by Pr\'{i}ncipe et al. in 2000. In this paper, we extend the classic CS divergence to quantify the closeness between two conditional distributions and show that the developed conditional CS divergence can be simply estimated by a kernel density estimator from given samples. We illustrate the advantages (e.g., the rigorous faithfulness guarantee, the lower computational complexity, the higher statistical power, and the much more flexibility in a wide range of applications) of our conditional CS divergence over previous proposals, such as the conditional KL divergence and the conditional maximum mean discrepancy. We also demonstrate the compelling performance of conditional CS divergence in two machine learning tasks related to time series data and sequential inference, namely the time series clustering and the uncertainty-guided exploration for sequential decision making.
Causal Recurrent Variational Autoencoder for Medical Time Series Generation
Jan 16, 2023Abstract:We propose causal recurrent variational autoencoder (CR-VAE), a novel generative model that is able to learn a Granger causal graph from a multivariate time series x and incorporates the underlying causal mechanism into its data generation process. Distinct to the classical recurrent VAEs, our CR-VAE uses a multi-head decoder, in which the $p$-th head is responsible for generating the $p$-th dimension of $\mathbf{x}$ (i.e., $\mathbf{x}^p$). By imposing a sparsity-inducing penalty on the weights (of the decoder) and encouraging specific sets of weights to be zero, our CR-VAE learns a sparse adjacency matrix that encodes causal relations between all pairs of variables. Thanks to this causal matrix, our decoder strictly obeys the underlying principles of Granger causality, thereby making the data generating process transparent. We develop a two-stage approach to train the overall objective. Empirically, we evaluate the behavior of our model in synthetic data and two real-world human brain datasets involving, respectively, the electroencephalography (EEG) signals and the functional magnetic resonance imaging (fMRI) data. Our model consistently outperforms state-of-the-art time series generative models both qualitatively and quantitatively. Moreover, it also discovers a faithful causal graph with similar or improved accuracy over existing Granger causality-based causal inference methods. Code of CR-VAE is publicly available at https://github.com/hongmingli1995/CR-VAE.
Deep Deterministic Independent Component Analysis for Hyperspectral Unmixing
Feb 15, 2022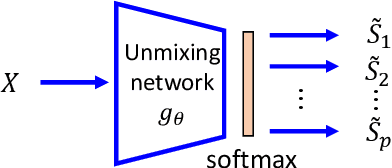
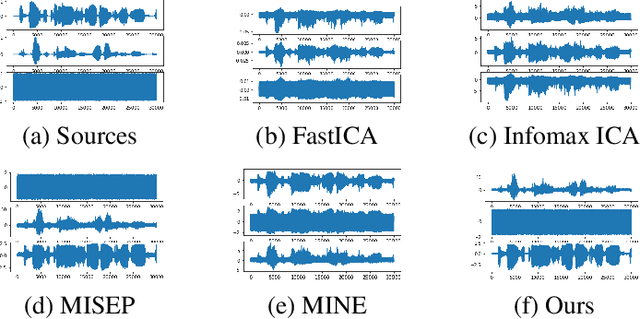
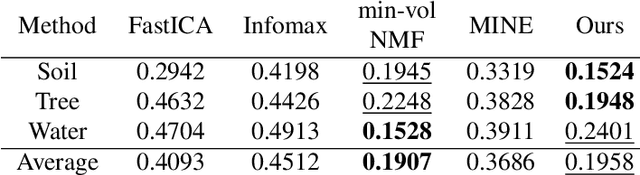
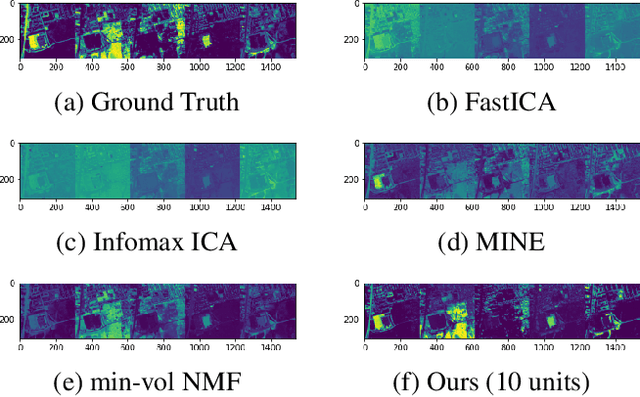
Abstract:We develop a new neural network based independent component analysis (ICA) method by directly minimizing the dependence amongst all extracted components. Using the matrix-based R{\'e}nyi's $\alpha$-order entropy functional, our network can be directly optimized by stochastic gradient descent (SGD), without any variational approximation or adversarial training. As a solid application, we evaluate our ICA in the problem of hyperspectral unmixing (HU) and refute a statement that "\emph{ICA does not play a role in unmixing hyperspectral data}", which was initially suggested by \cite{nascimento2005does}. Code and additional remarks of our DDICA is available at https://github.com/hongmingli1995/DDICA.
Unsupervised deep learning for individualized brain functional network identification
Dec 11, 2020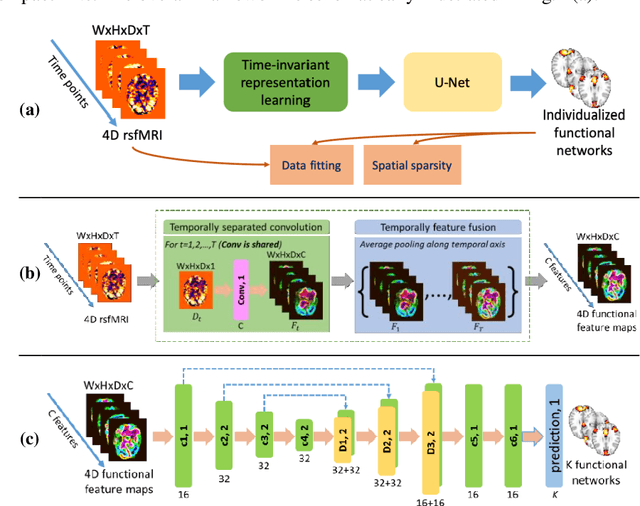
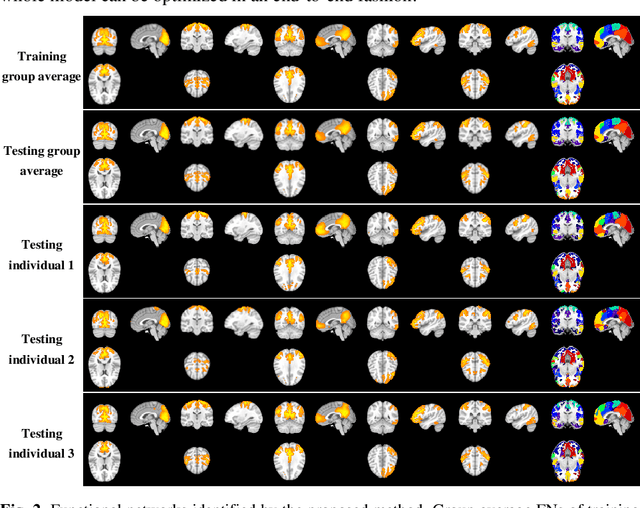
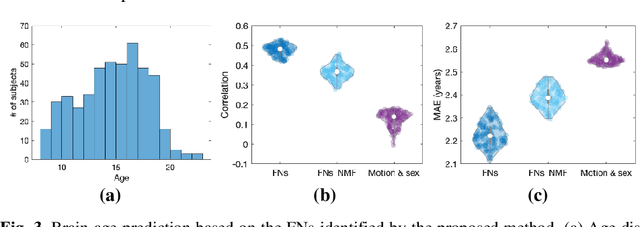
Abstract:A novel unsupervised deep learning method is developed to identify individual-specific large scale brain functional networks (FNs) from resting-state fMRI (rsfMRI) in an end-to-end learning fashion. Our method leverages deep Encoder-Decoder networks and conventional brain decomposition models to identify individual-specific FNs in an unsupervised learning framework and facilitate fast inference for new individuals with one forward pass of the deep network. Particularly, convolutional neural networks (CNNs) with an Encoder-Decoder architecture are adopted to identify individual-specific FNs from rsfMRI data by optimizing their data fitting and sparsity regularization terms that are commonly used in brain decomposition models. Moreover, a time-invariant representation learning module is designed to learn features invariant to temporal orders of time points of rsfMRI data. The proposed method has been validated based on a large rsfMRI dataset and experimental results have demonstrated that our method could obtain individual-specific FNs which are consistent with well-established FNs and are informative for predicting brain age, indicating that the individual-specific FNs identified truly captured the underlying variability of individualized functional neuroanatomy.
MDReg-Net: Multi-resolution diffeomorphic image registration using fully convolutional networks with deep self-supervision
Oct 04, 2020
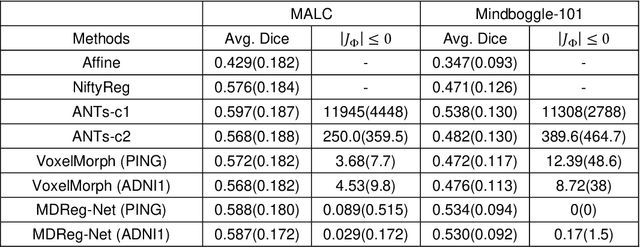


Abstract:We present a diffeomorphic image registration algorithm to learn spatial transformations between pairs of images to be registered using fully convolutional networks (FCNs) under a self-supervised learning setting. The network is trained to estimate diffeomorphic spatial transformations between pairs of images by maximizing an image-wise similarity metric between fixed and warped moving images, similar to conventional image registration algorithms. It is implemented in a multi-resolution image registration framework to optimize and learn spatial transformations at different image resolutions jointly and incrementally with deep self-supervision in order to better handle large deformation between images. A spatial Gaussian smoothing kernel is integrated with the FCNs to yield sufficiently smooth deformation fields to achieve diffeomorphic image registration. Particularly, spatial transformations learned at coarser resolutions are utilized to warp the moving image, which is subsequently used for learning incremental transformations at finer resolutions. This procedure proceeds recursively to the full image resolution and the accumulated transformations serve as the final transformation to warp the moving image at the finest resolution. Experimental results for registering high resolution 3D structural brain magnetic resonance (MR) images have demonstrated that image registration networks trained by our method obtain robust, diffeomorphic image registration results within seconds with improved accuracy compared with state-of-the-art image registration algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge