Robert Jenssen
Keypoint Counting Classifiers: Turning Vision Transformers into Self-Explainable Models Without Training
Dec 19, 2025
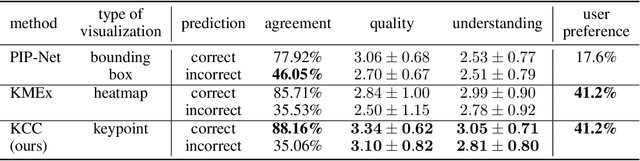

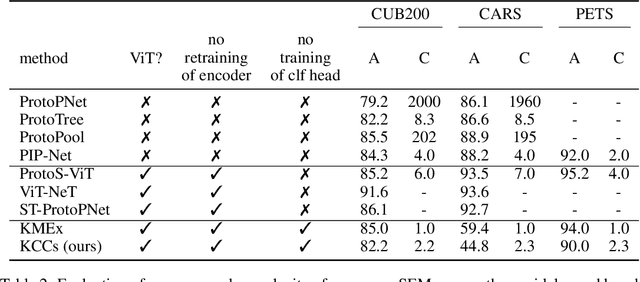
Abstract:Current approaches for designing self-explainable models (SEMs) require complicated training procedures and specific architectures which makes them impractical. With the advance of general purpose foundation models based on Vision Transformers (ViTs), this impracticability becomes even more problematic. Therefore, new methods are necessary to provide transparency and reliability to ViT-based foundation models. In this work, we present a new method for turning any well-trained ViT-based model into a SEM without retraining, which we call Keypoint Counting Classifiers (KCCs). Recent works have shown that ViTs can automatically identify matching keypoints between images with high precision, and we build on these results to create an easily interpretable decision process that is inherently visualizable in the input. We perform an extensive evaluation which show that KCCs improve the human-machine communication compared to recent baselines. We believe that KCCs constitute an important step towards making ViT-based foundation models more transparent and reliable.
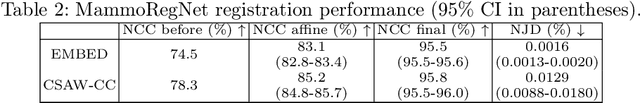
The Impact of Longitudinal Mammogram Alignment on Breast Cancer Risk Assessment
Nov 11, 2025Abstract:Regular mammography screening is crucial for early breast cancer detection. By leveraging deep learning-based risk models, screening intervals can be personalized, especially for high-risk individuals. While recent methods increasingly incorporate longitudinal information from prior mammograms, accurate spatial alignment across time points remains a key challenge. Misalignment can obscure meaningful tissue changes and degrade model performance. In this study, we provide insights into various alignment strategies, image-based registration, feature-level (representation space) alignment with and without regularization, and implicit alignment methods, for their effectiveness in longitudinal deep learning-based risk modeling. Using two large-scale mammography datasets, we assess each method across key metrics, including predictive accuracy, precision, recall, and deformation field quality. Our results show that image-based registration consistently outperforms the more recently favored feature-based and implicit approaches across all metrics, enabling more accurate, temporally consistent predictions and generating smooth, anatomically plausible deformation fields. Although regularizing the deformation field improves deformation quality, it reduces the risk prediction performance of feature-level alignment. Applying image-based deformation fields within the feature space yields the best risk prediction performance. These findings underscore the importance of image-based deformation fields for spatial alignment in longitudinal risk modeling, offering improved prediction accuracy and robustness. This approach has strong potential to enhance personalized screening and enable earlier interventions for high-risk individuals. The code is available at https://github.com/sot176/Mammogram_Alignment_Study_Risk_Prediction.git, allowing full reproducibility of the results.
A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging
Jul 03, 2025![Figure 1 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F5-Figure1-1.png&w=640&q=75)
![Figure 2 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F6-Figure2-1.png&w=640&q=75)
![Figure 3 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F7-Figure3-1.png&w=640&q=75)
![Figure 4 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F9-Figure4-1.png&w=640&q=75)
Abstract:Dynamic positron emission tomography (PET) and kinetic modeling are pivotal in advancing tracer development research in small animal studies. Accurate kinetic modeling requires precise input function estimation, traditionally achieved via arterial blood sampling. However, arterial cannulation in small animals like mice, involves intricate, time-consuming, and terminal procedures, precluding longitudinal studies. This work proposes a non-invasive, fully convolutional deep learning-based approach (FC-DLIF) to predict input functions directly from PET imaging, potentially eliminating the need for blood sampling in dynamic small-animal PET. The proposed FC-DLIF model includes a spatial feature extractor acting on the volumetric time frames of the PET sequence, extracting spatial features. These are subsequently further processed in a temporal feature extractor that predicts the arterial input function. The proposed approach is trained and evaluated using images and arterial blood curves from [$^{18}$F]FDG data using cross validation. Further, the model applicability is evaluated on imaging data and arterial blood curves collected using two additional radiotracers ([$^{18}$F]FDOPA, and [$^{68}$Ga]PSMA). The model was further evaluated on data truncated and shifted in time, to simulate shorter, and shifted, PET scans. The proposed FC-DLIF model reliably predicts the arterial input function with respect to mean squared error and correlation. Furthermore, the FC-DLIF model is able to predict the arterial input function even from truncated and shifted samples. The model fails to predict the AIF from samples collected using different radiotracers, as these are not represented in the training data. Our deep learning-based input function offers a non-invasive and reliable alternative to arterial blood sampling, proving robust and flexible to temporal shifts and different scan durations.
Reconsidering Explicit Longitudinal Mammography Alignment for Enhanced Breast Cancer Risk Prediction
Jun 24, 2025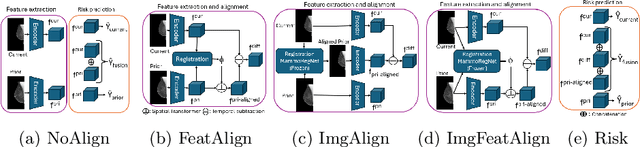
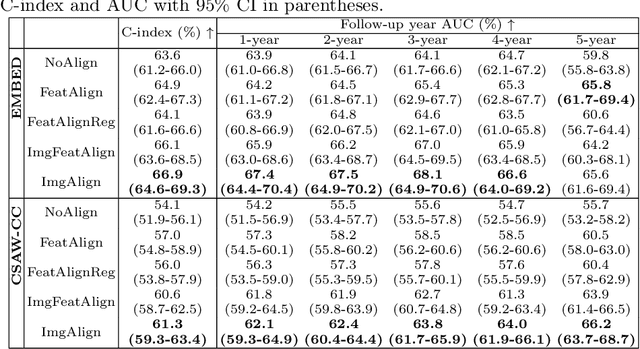
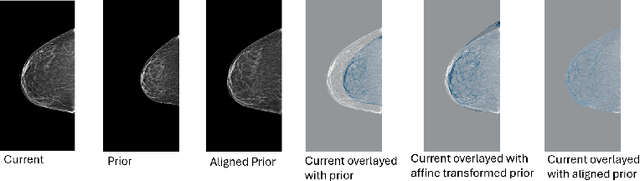
Abstract:Regular mammography screening is essential for early breast cancer detection. Deep learning-based risk prediction methods have sparked interest to adjust screening intervals for high-risk groups. While early methods focused only on current mammograms, recent approaches leverage the temporal aspect of screenings to track breast tissue changes over time, requiring spatial alignment across different time points. Two main strategies for this have emerged: explicit feature alignment through deformable registration and implicit learned alignment using techniques like transformers, with the former providing more control. However, the optimal approach for explicit alignment in mammography remains underexplored. In this study, we provide insights into where explicit alignment should occur (input space vs. representation space) and if alignment and risk prediction should be jointly optimized. We demonstrate that jointly learning explicit alignment in representation space while optimizing risk estimation performance, as done in the current state-of-the-art approach, results in a trade-off between alignment quality and predictive performance and show that image-level alignment is superior to representation-level alignment, leading to better deformation field quality and enhanced risk prediction accuracy. The code is available at https://github.com/sot176/Longitudinal_Mammogram_Alignment.git.
Aggregation of Dependent Expert Distributions in Multimodal Variational Autoencoders
May 02, 2025Abstract:Multimodal learning with variational autoencoders (VAEs) requires estimating joint distributions to evaluate the evidence lower bound (ELBO). Current methods, the product and mixture of experts, aggregate single-modality distributions assuming independence for simplicity, which is an overoptimistic assumption. This research introduces a novel methodology for aggregating single-modality distributions by exploiting the principle of consensus of dependent experts (CoDE), which circumvents the aforementioned assumption. Utilizing the CoDE method, we propose a novel ELBO that approximates the joint likelihood of the multimodal data by learning the contribution of each subset of modalities. The resulting CoDE-VAE model demonstrates better performance in terms of balancing the trade-off between generative coherence and generative quality, as well as generating more precise log-likelihood estimations. CoDE-VAE further minimizes the generative quality gap as the number of modalities increases. In certain cases, it reaches a generative quality similar to that of unimodal VAEs, which is a desirable property that is lacking in most current methods. Finally, the classification accuracy achieved by CoDE-VAE is comparable to that of state-of-the-art multimodal VAE models.
From Colors to Classes: Emergence of Concepts in Vision Transformers
Mar 31, 2025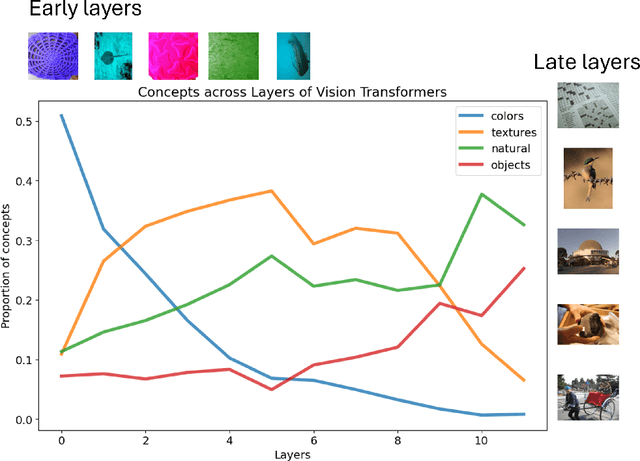
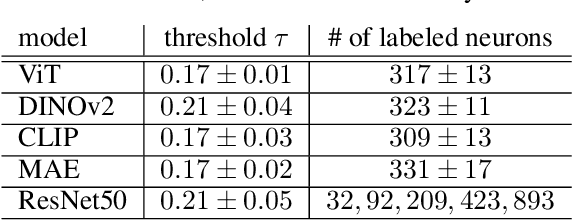
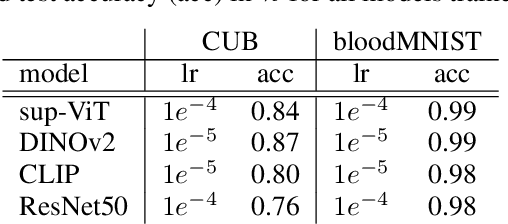
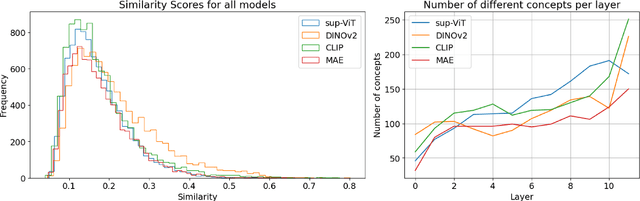
Abstract:Vision Transformers (ViTs) are increasingly utilized in various computer vision tasks due to their powerful representation capabilities. However, it remains understudied how ViTs process information layer by layer. Numerous studies have shown that convolutional neural networks (CNNs) extract features of increasing complexity throughout their layers, which is crucial for tasks like domain adaptation and transfer learning. ViTs, lacking the same inductive biases as CNNs, can potentially learn global dependencies from the first layers due to their attention mechanisms. Given the increasing importance of ViTs in computer vision, there is a need to improve the layer-wise understanding of ViTs. In this work, we present a novel, layer-wise analysis of concepts encoded in state-of-the-art ViTs using neuron labeling. Our findings reveal that ViTs encode concepts with increasing complexity throughout the network. Early layers primarily encode basic features such as colors and textures, while later layers represent more specific classes, including objects and animals. As the complexity of encoded concepts increases, the number of concepts represented in each layer also rises, reflecting a more diverse and specific set of features. Additionally, different pretraining strategies influence the quantity and category of encoded concepts, with finetuning to specific downstream tasks generally reducing the number of encoded concepts and shifting the concepts to more relevant categories.
Natural Language Processing for Electronic Health Records in Scandinavian Languages: Norwegian, Swedish, and Danish
Mar 24, 2025



Abstract:Background: Clinical natural language processing (NLP) refers to the use of computational methods for extracting, processing, and analyzing unstructured clinical text data, and holds a huge potential to transform healthcare in various clinical tasks. Objective: The study aims to perform a systematic review to comprehensively assess and analyze the state-of-the-art NLP methods for the mainland Scandinavian clinical text. Method: A literature search was conducted in various online databases including PubMed, ScienceDirect, Google Scholar, ACM digital library, and IEEE Xplore between December 2022 and February 2024. Further, relevant references to the included articles were also used to solidify our search. The final pool includes articles that conducted clinical NLP in the mainland Scandinavian languages and were published in English between 2010 and 2024. Results: Out of the 113 articles, 18% (n=21) focus on Norwegian clinical text, 64% (n=72) on Swedish, 10% (n=11) on Danish, and 8% (n=9) focus on more than one language. Generally, the review identified positive developments across the region despite some observable gaps and disparities between the languages. There are substantial disparities in the level of adoption of transformer-based models. In essential tasks such as de-identification, there is significantly less research activity focusing on Norwegian and Danish compared to Swedish text. Further, the review identified a low level of sharing resources such as data, experimentation code, pre-trained models, and rate of adaptation and transfer learning in the region. Conclusion: The review presented a comprehensive assessment of the state-of-the-art Clinical NLP for electronic health records (EHR) text in mainland Scandinavian languages and, highlighted the potential barriers and challenges that hinder the rapid advancement of the field in the region.
REPEAT: Improving Uncertainty Estimation in Representation Learning Explainability
Dec 11, 2024Abstract:Incorporating uncertainty is crucial to provide trustworthy explanations of deep learning models. Recent works have demonstrated how uncertainty modeling can be particularly important in the unsupervised field of representation learning explainable artificial intelligence (R-XAI). Current R-XAI methods provide uncertainty by measuring variability in the importance score. However, they fail to provide meaningful estimates of whether a pixel is certainly important or not. In this work, we propose a new R-XAI method called REPEAT that addresses the key question of whether or not a pixel is \textit{certainly} important. REPEAT leverages the stochasticity of current R-XAI methods to produce multiple estimates of importance, thus considering each pixel in an image as a Bernoulli random variable that is either important or unimportant. From these Bernoulli random variables we can directly estimate the importance of a pixel and its associated certainty, thus enabling users to determine certainty in pixel importance. Our extensive evaluation shows that REPEAT gives certainty estimates that are more intuitive, better at detecting out-of-distribution data, and more concise.
Explaining time series models using frequency masking
Jun 19, 2024Abstract:Time series data is fundamentally important for describing many critical domains such as healthcare, finance, and climate, where explainable models are necessary for safe automated decision-making. To develop eXplainable AI (XAI) in these domains therefore implies explaining salient information in the time series. Current methods for obtaining saliency maps assumes localized information in the raw input space. In this paper, we argue that the salient information of a number of time series is more likely to be localized in the frequency domain. We propose FreqRISE, which uses masking based methods to produce explanations in the frequency and time-frequency domain, which shows the best performance across a number of tasks.
Generalized Cauchy-Schwarz Divergence and Its Deep Learning Applications
May 07, 2024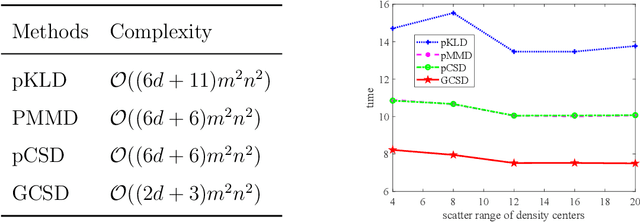
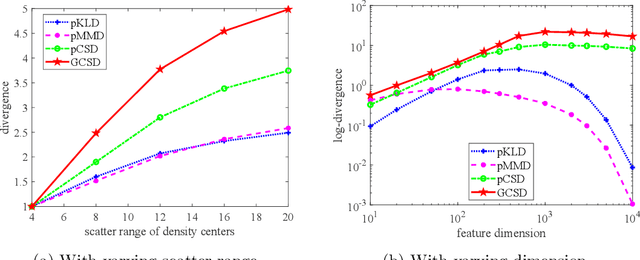
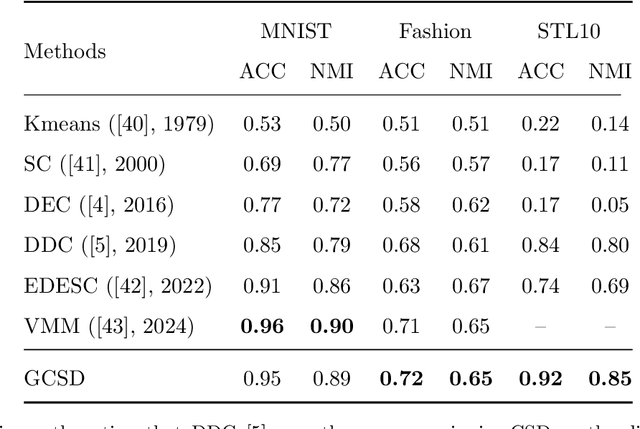
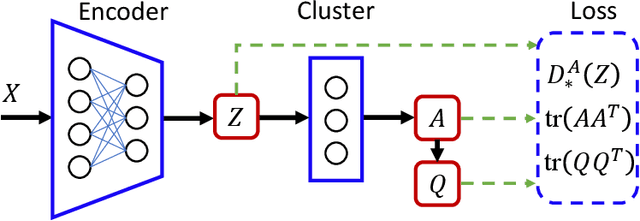
Abstract:Divergence measures play a central role in machine learning and become increasingly essential in deep learning. However, valid and computationally efficient divergence measures for multiple (more than two) distributions are scarcely investigated. This becomes particularly crucial in areas where the simultaneous management of multiple distributions is both unavoidable and essential. Examples include clustering, multi-source domain adaptation or generalization, and multi-view learning, among others. Although calculating the mean of pairwise distances between any two distributions serves as a common way to quantify the total divergence among multiple distributions, it is crucial to acknowledge that this approach is not straightforward and requires significant computational resources. In this study, we introduce a new divergence measure for multiple distributions named the generalized Cauchy-Schwarz divergence (GCSD), which is inspired by the classic Cauchy-Schwarz divergence. Additionally, we provide a closed-form sample estimator based on kernel density estimation, making it convenient and straightforward to use in various machine-learning applications. Finally, we apply the proposed GCSD to two challenging machine learning tasks, namely deep learning-based clustering and the problem of multi-source domain adaptation. The experimental results showcase the impressive performance of GCSD in both tasks, highlighting its potential application in machine-learning areas that involve quantifying multiple distributions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge