Elisabeth Wetzer
Keypoint Counting Classifiers: Turning Vision Transformers into Self-Explainable Models Without Training
Dec 19, 2025
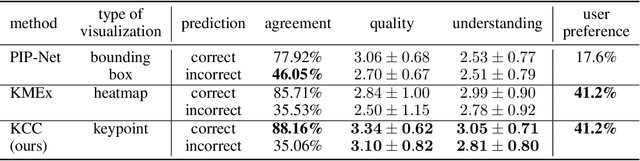

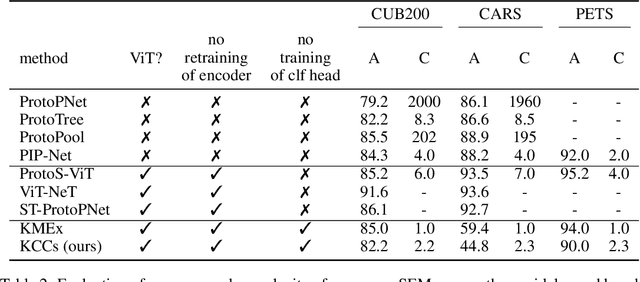
Abstract:Current approaches for designing self-explainable models (SEMs) require complicated training procedures and specific architectures which makes them impractical. With the advance of general purpose foundation models based on Vision Transformers (ViTs), this impracticability becomes even more problematic. Therefore, new methods are necessary to provide transparency and reliability to ViT-based foundation models. In this work, we present a new method for turning any well-trained ViT-based model into a SEM without retraining, which we call Keypoint Counting Classifiers (KCCs). Recent works have shown that ViTs can automatically identify matching keypoints between images with high precision, and we build on these results to create an easily interpretable decision process that is inherently visualizable in the input. We perform an extensive evaluation which show that KCCs improve the human-machine communication compared to recent baselines. We believe that KCCs constitute an important step towards making ViT-based foundation models more transparent and reliable.
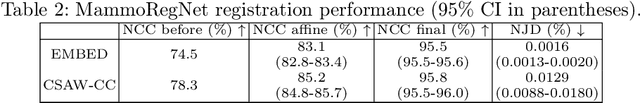
The Impact of Longitudinal Mammogram Alignment on Breast Cancer Risk Assessment
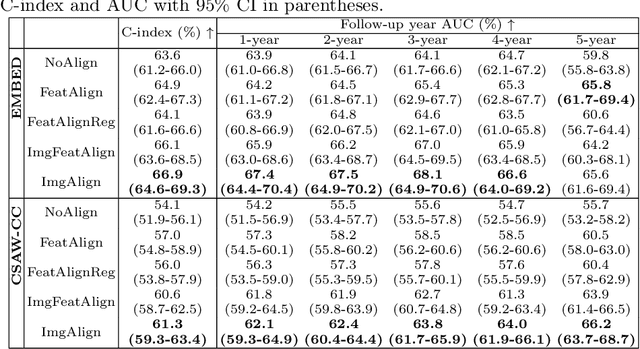
Nov 11, 2025Abstract:Regular mammography screening is crucial for early breast cancer detection. By leveraging deep learning-based risk models, screening intervals can be personalized, especially for high-risk individuals. While recent methods increasingly incorporate longitudinal information from prior mammograms, accurate spatial alignment across time points remains a key challenge. Misalignment can obscure meaningful tissue changes and degrade model performance. In this study, we provide insights into various alignment strategies, image-based registration, feature-level (representation space) alignment with and without regularization, and implicit alignment methods, for their effectiveness in longitudinal deep learning-based risk modeling. Using two large-scale mammography datasets, we assess each method across key metrics, including predictive accuracy, precision, recall, and deformation field quality. Our results show that image-based registration consistently outperforms the more recently favored feature-based and implicit approaches across all metrics, enabling more accurate, temporally consistent predictions and generating smooth, anatomically plausible deformation fields. Although regularizing the deformation field improves deformation quality, it reduces the risk prediction performance of feature-level alignment. Applying image-based deformation fields within the feature space yields the best risk prediction performance. These findings underscore the importance of image-based deformation fields for spatial alignment in longitudinal risk modeling, offering improved prediction accuracy and robustness. This approach has strong potential to enhance personalized screening and enable earlier interventions for high-risk individuals. The code is available at https://github.com/sot176/Mammogram_Alignment_Study_Risk_Prediction.git, allowing full reproducibility of the results.
A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging
Jul 03, 2025![Figure 1 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F5-Figure1-1.png&w=640&q=75)
![Figure 2 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F6-Figure2-1.png&w=640&q=75)
![Figure 3 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F7-Figure3-1.png&w=640&q=75)
![Figure 4 for A robust and versatile deep learning model for prediction of the arterial input function in dynamic small animal $\left[^{18}\text{F}\right]$FDG PET imaging](/_next/image?url=https%3A%2F%2Ffigures.semanticscholar.org%2Faeedac3751b9e214478a1237ed9043e09178057f%2F9-Figure4-1.png&w=640&q=75)
Abstract:Dynamic positron emission tomography (PET) and kinetic modeling are pivotal in advancing tracer development research in small animal studies. Accurate kinetic modeling requires precise input function estimation, traditionally achieved via arterial blood sampling. However, arterial cannulation in small animals like mice, involves intricate, time-consuming, and terminal procedures, precluding longitudinal studies. This work proposes a non-invasive, fully convolutional deep learning-based approach (FC-DLIF) to predict input functions directly from PET imaging, potentially eliminating the need for blood sampling in dynamic small-animal PET. The proposed FC-DLIF model includes a spatial feature extractor acting on the volumetric time frames of the PET sequence, extracting spatial features. These are subsequently further processed in a temporal feature extractor that predicts the arterial input function. The proposed approach is trained and evaluated using images and arterial blood curves from [$^{18}$F]FDG data using cross validation. Further, the model applicability is evaluated on imaging data and arterial blood curves collected using two additional radiotracers ([$^{18}$F]FDOPA, and [$^{68}$Ga]PSMA). The model was further evaluated on data truncated and shifted in time, to simulate shorter, and shifted, PET scans. The proposed FC-DLIF model reliably predicts the arterial input function with respect to mean squared error and correlation. Furthermore, the FC-DLIF model is able to predict the arterial input function even from truncated and shifted samples. The model fails to predict the AIF from samples collected using different radiotracers, as these are not represented in the training data. Our deep learning-based input function offers a non-invasive and reliable alternative to arterial blood sampling, proving robust and flexible to temporal shifts and different scan durations.
Reconsidering Explicit Longitudinal Mammography Alignment for Enhanced Breast Cancer Risk Prediction
Jun 24, 2025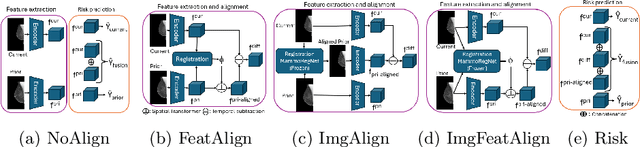
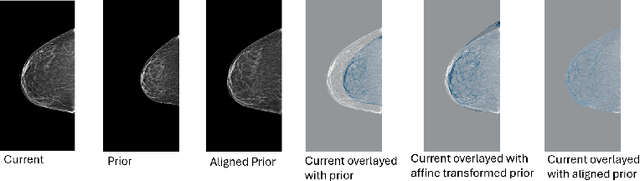
Abstract:Regular mammography screening is essential for early breast cancer detection. Deep learning-based risk prediction methods have sparked interest to adjust screening intervals for high-risk groups. While early methods focused only on current mammograms, recent approaches leverage the temporal aspect of screenings to track breast tissue changes over time, requiring spatial alignment across different time points. Two main strategies for this have emerged: explicit feature alignment through deformable registration and implicit learned alignment using techniques like transformers, with the former providing more control. However, the optimal approach for explicit alignment in mammography remains underexplored. In this study, we provide insights into where explicit alignment should occur (input space vs. representation space) and if alignment and risk prediction should be jointly optimized. We demonstrate that jointly learning explicit alignment in representation space while optimizing risk estimation performance, as done in the current state-of-the-art approach, results in a trade-off between alignment quality and predictive performance and show that image-level alignment is superior to representation-level alignment, leading to better deformation field quality and enhanced risk prediction accuracy. The code is available at https://github.com/sot176/Longitudinal_Mammogram_Alignment.git.
Can representation learning for multimodal image registration be improved by supervision of intermediate layers?
Mar 01, 2023Abstract:Multimodal imaging and correlative analysis typically require image alignment. Contrastive learning can generate representations of multimodal images, reducing the challenging task of multimodal image registration to a monomodal one. Previously, additional supervision on intermediate layers in contrastive learning has improved biomedical image classification. We evaluate if a similar approach improves representations learned for registration to boost registration performance. We explore three approaches to add contrastive supervision to the latent features of the bottleneck layer in the U-Nets encoding the multimodal images and evaluate three different critic functions. Our results show that representations learned without additional supervision on latent features perform best in the downstream task of registration on two public biomedical datasets. We investigate the performance drop by exploiting recent insights in contrastive learning in classification and self-supervised learning. We visualize the spatial relations of the learned representations by means of multidimensional scaling, and show that additional supervision on the bottleneck layer can lead to partial dimensional collapse of the intermediate embedding space.
Cross-Modality Sub-Image Retrieval using Contrastive Multimodal Image Representations
Jan 10, 2022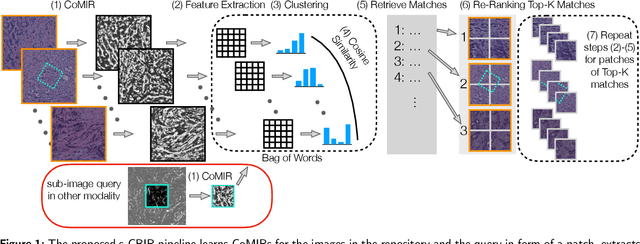
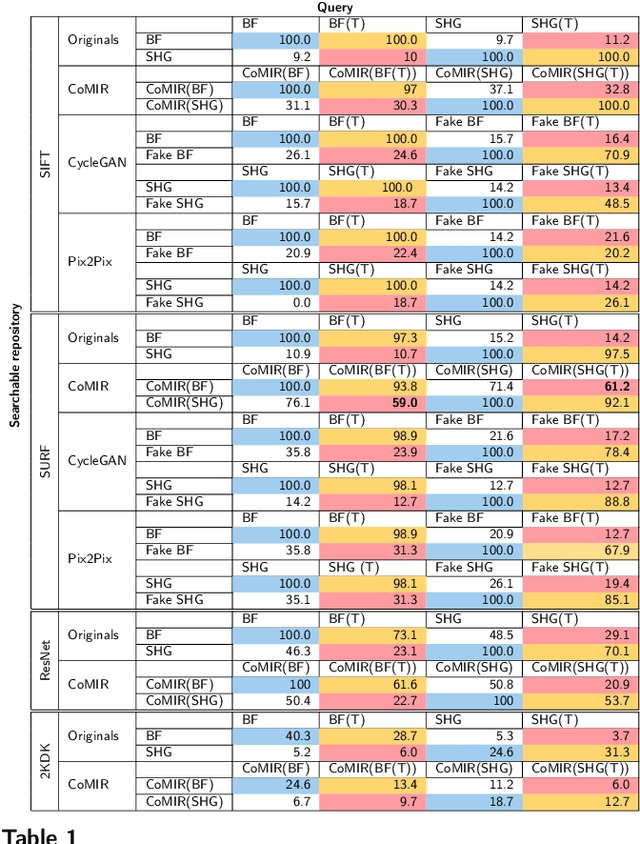
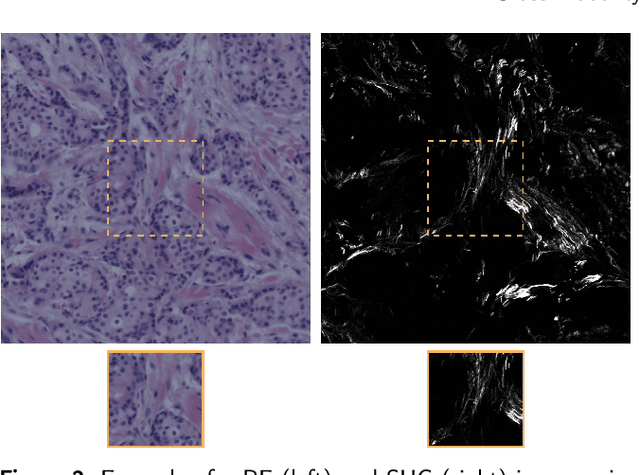
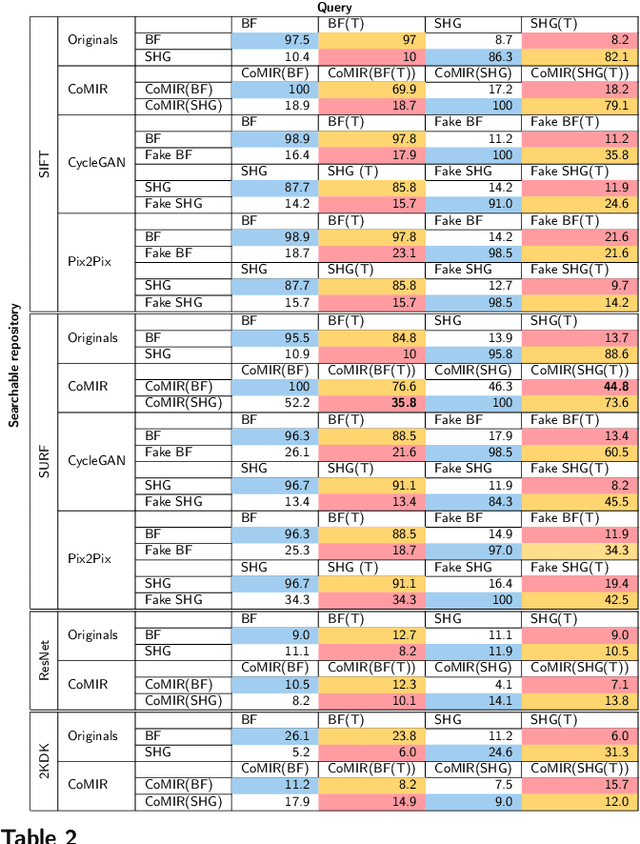
Abstract:In tissue characterization and cancer diagnostics, multimodal imaging has emerged as a powerful technique. Thanks to computational advances, large datasets can be exploited to improve diagnosis and discover patterns in pathologies. However, this requires efficient and scalable image retrieval methods. Cross-modality image retrieval is particularly demanding, as images of the same content captured in different modalities may display little common information. We propose a content-based image retrieval system (CBIR) for reverse (sub-)image search to retrieve microscopy images in one modality given a corresponding image captured by a different modality, where images are not aligned and share only few structures. We propose to combine deep learning to generate representations which embed both modalities in a common space, with classic, fast, and robust feature extractors (SIFT, SURF) to create a bag-of-words model for efficient and reliable retrieval. Our application-independent approach shows promising results on a publicly available dataset of brightfield and second harmonic generation microscopy images. We obtain 75.4% and 83.6% top-10 retrieval success for retrieval in one or the other direction. Our proposed method significantly outperforms both direct retrieval of the original multimodal (sub-)images, as well as their corresponding generative adversarial network (GAN)-based image-to-image translations. We establish that the proposed method performs better in comparison with a recent sub-image retrieval toolkit, GAN-based image-to-image translations, and learnt feature extractors for the downstream task of cross-modal image retrieval. We highlight the shortcomings of the latter methods and observe the importance of equivariance and invariance properties of the learnt representations and feature extractors in the CBIR pipeline. Code will be available at github.com/MIDA-group.
CoMIR: Contrastive Multimodal Image Representation for Registration
Jun 11, 2020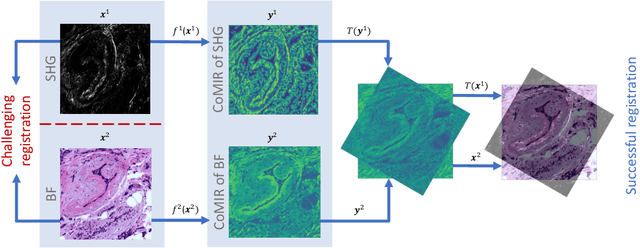

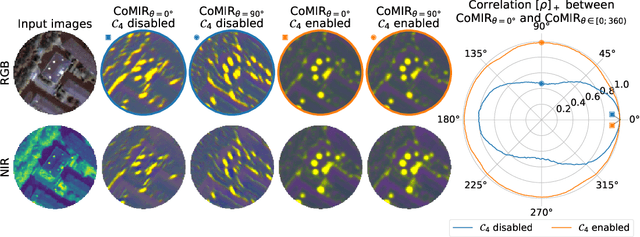
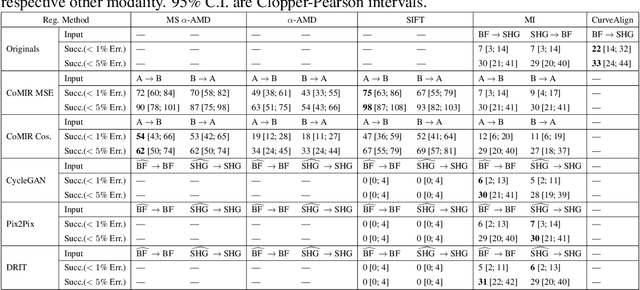
Abstract:We propose contrastive coding to learn shared, dense image representations, referred to as CoMIRs (Contrastive Multimodal Image Representations). CoMIRs enable the registration of multimodal images where existing registration methods often fail due to a lack of sufficiently similar image structures. CoMIRs reduce the multimodal registration problem to a monomodal one in which general intensity-based, as well as feature-based, registration algorithms can be applied. The method involves training one neural network per modality on aligned images, using a contrastive loss based on noise-contrastive estimation (InfoNCE). Unlike other contrastive coding methods, used for e.g. classification, our approach generates image-like representations that contain the information shared between modalities. We introduce a novel, hyperparameter-free modification to InfoNCE, to enforce rotational equivariance of the learnt representations, a property essential to the registration task. We assess the extent of achieved rotational equivariance and the stability of the representations with respect to weight initialization, training set, and hyperparameter settings, on a remote sensing dataset of RGB and near-infrared images. We evaluate the learnt representations through registration of a biomedical dataset of bright-field and second-harmonic generation microscopy images; two modalities with very little apparent correlation. The proposed approach based on CoMIRs significantly outperforms registration of representations created by GAN-based image-to-image translation, as well as a state-of-the-art, application-specific method which takes additional knowledge about the data into account. Code is available at: https://github.com/dqiamsdoayehccdvulyy/CoMIR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge