Johan Öfverstedt
Towards prediction of morphological heart age from computed tomography angiography
Apr 22, 2025Abstract:Age prediction from medical images or other health-related non-imaging data is an important approach to data-driven aging research, providing knowledge of how much information a specific tissue or organ carries about the chronological age of the individual. In this work, we studied the prediction of age from computed tomography angiography (CTA) images, which provide detailed representations of the heart morphology, with the goals of (i) studying the relationship between morphology and aging, and (ii) developing a novel \emph{morphological heart age} biomarker. We applied an image registration-based method that standardizes the images from the whole cohort into a single space. We then extracted supervoxels (using unsupervised segmentation), and corresponding robust features of density and local volume, which provide a detailed representation of the heart morphology while being robust to registration errors. Machine learning models are then trained to fit regression models from these features to the chronological age. We applied the method to a subset of the images from the Swedish CArdioPulomonary bioImage Study (SCAPIS) dataset, consisting of 721 females and 666 males. We observe a mean absolute error of $2.74$ years for females and $2.77$ years for males. The predictions from different sub-regions of interest were observed to be more highly correlated with the predictions from the whole heart, compared to the chronological age, revealing a high consistency in the predictions from morphology. Saliency analysis was also performed on the prediction models to study what regions are associated positively and negatively with the predicted age. This resulted in detailed association maps where the density and volume of known, as well as some novel sub-regions of interest, are determined to be important. The saliency analysis aids in the interpretability of the models and their predictions.
A method for supervoxel-wise association studies of age and other non-imaging variables from coronary computed tomography angiograms
May 13, 2024Abstract:The study of associations between an individual's age and imaging and non-imaging data is an active research area that attempts to aid understanding of the effects and patterns of aging. In this work we have conducted a supervoxel-wise association study between both volumetric and tissue density features in coronary computed tomography angiograms and the chronological age of a subject, to understand the localized changes in morphology and tissue density with age. To enable a supervoxel-wise study of volume and tissue density, we developed a novel method based on image segmentation, inter-subject image registration, and robust supervoxel-based correlation analysis, to achieve a statistical association study between the images and age. We evaluate the registration methodology in terms of the Dice coefficient for the heart chambers and myocardium, and the inverse consistency of the transformations, showing that the method works well in most cases with high overlap and inverse consistency. In a sex-stratified study conducted on a subset of $n=1388$ images from the SCAPIS study, the supervoxel-wise analysis was able to find localized associations with age outside of the commonly segmented and analyzed sub-regions, and several substantial differences between the sexes in association of age and volume.
Cross-Sim-NGF: FFT-Based Global Rigid Multimodal Alignment of Image Volumes using Normalized Gradient Fields
Oct 19, 2021



Abstract:Multimodal image alignment involves finding spatial correspondences between volumes varying in appearance and structure. Automated alignment methods are often based on local optimization that can be highly sensitive to their initialization. We propose a global optimization method for rigid multimodal 3D image alignment, based on a novel efficient algorithm for computing similarity of normalized gradient fields (NGF) in the frequency domain. We validate the method experimentally on a dataset comprised of 20 brain volumes acquired in four modalities (T1w, Flair, CT, [18F] FDG PET), synthetically displaced with known transformations. The proposed method exhibits excellent performance on all six possible modality combinations, and outperforms all four reference methods by a large margin. The method is fast; a 3.4Mvoxel global rigid alignment requires approximately 40 seconds of computation, and the proposed algorithm outperforms a direct algorithm for the same task by more than three orders of magnitude. Open-source implementation is provided.
Fast computation of mutual information in the frequency domain with applications to global multimodal image alignment
Jun 28, 2021



Abstract:Multimodal image alignment is the process of finding spatial correspondences between images formed by different imaging techniques or under different conditions, to facilitate heterogeneous data fusion and correlative analysis. The information-theoretic concept of mutual information (MI) is widely used as a similarity measure to guide multimodal alignment processes, where most works have focused on local maximization of MI that typically works well only for small displacements; this points to a need for global maximization of MI, which has previously been computationally infeasible due to the high run-time complexity of existing algorithms. We propose an efficient algorithm for computing MI for all discrete displacements (formalized as the cross-mutual information function (CMIF)), which is based on cross-correlation computed in the frequency domain. We show that the algorithm is equivalent to a direct method while asymptotically superior in terms of run-time. Furthermore, we propose a method for multimodal image alignment for transformation models with few degrees of freedom (e.g. rigid) based on the proposed CMIF-algorithm. We evaluate the efficacy of the proposed method on three distinct benchmark datasets, of aerial images, cytological images, and histological images, and we observe excellent success-rates (in recovering known rigid transformations), overall outperforming alternative methods, including local optimization of MI as well as several recent deep learning-based approaches. We also evaluate the run-times of a GPU implementation of the proposed algorithm and observe speed-ups from 100 to more than 10,000 times for realistic image sizes compared to a GPU implementation of a direct method. Code is shared as open-source at \url{github.com/MIDA-group/globalign}.
Is Image-to-Image Translation the Panacea for Multimodal Image Registration? A Comparative Study
Mar 30, 2021



Abstract:Despite current advancement in the field of biomedical image processing, propelled by the deep learning revolution, multimodal image registration, due to its several challenges, is still often performed manually by specialists. The recent success of image-to-image (I2I) translation in computer vision applications and its growing use in biomedical areas provide a tempting possibility of transforming the multimodal registration problem into a, potentially easier, monomodal one. We conduct an empirical study of the applicability of modern I2I translation methods for the task of multimodal biomedical image registration. We compare the performance of four Generative Adversarial Network (GAN)-based methods and one contrastive representation learning method, subsequently combined with two representative monomodal registration methods, to judge the effectiveness of modality translation for multimodal image registration. We evaluate these method combinations on three publicly available multimodal datasets of increasing difficulty, and compare with the performance of registration by Mutual Information maximisation and one modern data-specific multimodal registration method. Our results suggest that, although I2I translation may be helpful when the modalities to register are clearly correlated, registration of modalities which express distinctly different properties of the sample are not well handled by the I2I translation approach. When less information is shared between the modalities, the I2I translation methods struggle to provide good predictions, which impairs the registration performance. The evaluated representation learning method, which aims to find an in-between representation, manages better, and so does the Mutual Information maximisation approach. We share our complete experimental setup as open-source (https://github.com/Noodles-321/Registration).
INSPIRE: Intensity and Spatial Information-Based Deformable Image Registration
Dec 14, 2020



Abstract:We present INSPIRE, a top-performing general-purpose method for deformable image registration. INSPIRE extends our existing symmetric registration framework based on distances combining intensity and spatial information to an elastic B-splines based transformation model. We also present several theoretical and algorithmic improvements which provide high computational efficiency and thereby applicability of the framework in a wide range of real scenarios. We show that the proposed method delivers both highly accurate as well as stable and robust registration results. We evaluate the method on a synthetic dataset created from retinal images, consisting of thin networks of vessels, where INSPIRE exhibits excellent performance, substantially outperforming the reference methods. We also evaluate the method on four benchmark datasets of 3D images of brains, for a total of 2088 pairwise registrations; a comparison with 15 other state-of-the-art methods reveals that INSPIRE provides the best overall performance. Code is available at github.com/MIDA-group/inspire.
CoMIR: Contrastive Multimodal Image Representation for Registration
Jun 11, 2020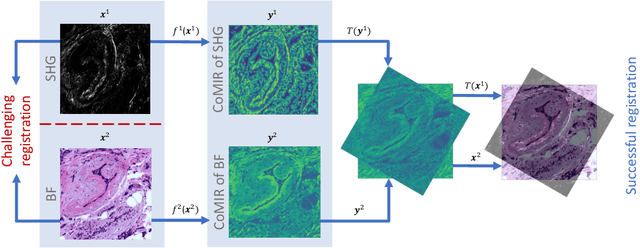

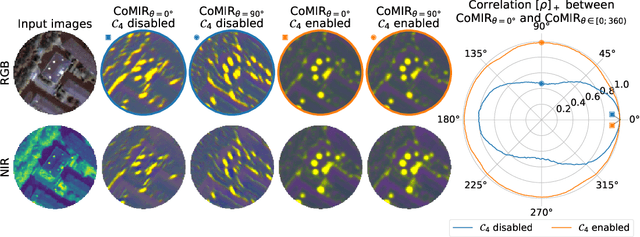
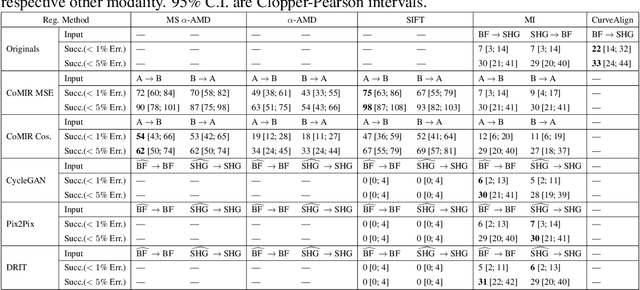
Abstract:We propose contrastive coding to learn shared, dense image representations, referred to as CoMIRs (Contrastive Multimodal Image Representations). CoMIRs enable the registration of multimodal images where existing registration methods often fail due to a lack of sufficiently similar image structures. CoMIRs reduce the multimodal registration problem to a monomodal one in which general intensity-based, as well as feature-based, registration algorithms can be applied. The method involves training one neural network per modality on aligned images, using a contrastive loss based on noise-contrastive estimation (InfoNCE). Unlike other contrastive coding methods, used for e.g. classification, our approach generates image-like representations that contain the information shared between modalities. We introduce a novel, hyperparameter-free modification to InfoNCE, to enforce rotational equivariance of the learnt representations, a property essential to the registration task. We assess the extent of achieved rotational equivariance and the stability of the representations with respect to weight initialization, training set, and hyperparameter settings, on a remote sensing dataset of RGB and near-infrared images. We evaluate the learnt representations through registration of a biomedical dataset of bright-field and second-harmonic generation microscopy images; two modalities with very little apparent correlation. The proposed approach based on CoMIRs significantly outperforms registration of representations created by GAN-based image-to-image translation, as well as a state-of-the-art, application-specific method which takes additional knowledge about the data into account. Code is available at: https://github.com/dqiamsdoayehccdvulyy/CoMIR.
Stochastic Distance Transform
Oct 18, 2018



Abstract:The distance transform (DT) and its many variations are ubiquitous tools for image processing and analysis. In many imaging scenarios, the images of interest are corrupted by noise. This has a strong negative impact on the accuracy of the DT, which is highly sensitive to spurious noise points. In this study, we consider images represented as discrete random sets and observe statistics of DT computed on such representations. We, thus, define a stochastic distance transform (SDT), which has an adjustable robustness to noise. Both a stochastic Monte Carlo method and a deterministic method for computing the SDT are proposed and compared. Through a series of empirical tests, we demonstrate that the SDT is effective not only in improving the accuracy of the computed distances in the presence of noise, but also in improving the performance of template matching and watershed segmentation of partially overlapping objects, which are examples of typical applications where DTs are utilized.
Fast and Robust Symmetric Image Registration Based on Intensity and Spatial Information
Jul 30, 2018



Abstract:Intensity-based image registration approaches rely on similarity measures to guide the search for geometric correspondences with high affinity between images. The properties of the used measure are vital for the robustness and accuracy of the registration. In this study a symmetric, intensity interpolation-free, affine registration framework based on a combination of intensity and spatial information is proposed. The excellent performance of the framework is demonstrated on a combination of synthetic tests, recovering known transformations in the presence of noise, and real applications in biomedical and medical image registration, for both 2D and 3D images. The method exhibits greater robustness and higher accuracy than similarity measures in common use, when inserted into a standard gradient-based registration framework available as part of the open source Insight Segmentation and Registration Toolkit (ITK). The method is also empirically shown to have a low computational cost, making it practical for real applications. Source code is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge