Dar-In Tai
Accurate and Generalizable Quantitative Scoring of Liver Steatosis from Ultrasound Images via Scalable Deep Learning
Oct 12, 2021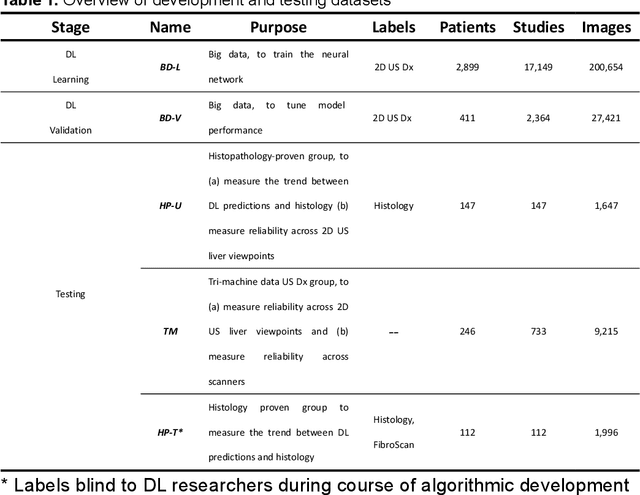


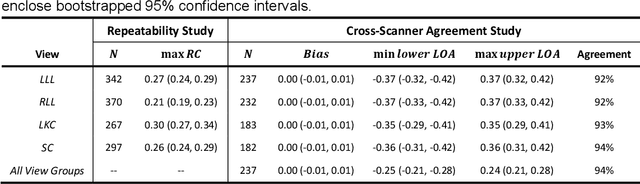
Abstract:Background & Aims: Hepatic steatosis is a major cause of chronic liver disease. 2D ultrasound is the most widely used non-invasive tool for screening and monitoring, but associated diagnoses are highly subjective. We developed a scalable deep learning (DL) algorithm for quantitative scoring of liver steatosis from 2D ultrasound images. Approach & Results: Using retrospectively collected multi-view ultrasound data from 3,310 patients, 19,513 studies, and 228,075 images, we trained a DL algorithm to diagnose steatosis stages (healthy, mild, moderate, or severe) from ultrasound diagnoses. Performance was validated on two multi-scanner unblinded and blinded (initially to DL developer) histology-proven cohorts (147 and 112 patients) with histopathology fatty cell percentage diagnoses, and a subset with FibroScan diagnoses. We also quantified reliability across scanners and viewpoints. Results were evaluated using Bland-Altman and receiver operating characteristic (ROC) analysis. The DL algorithm demonstrates repeatable measurements with a moderate number of images (3 for each viewpoint) and high agreement across 3 premium ultrasound scanners. High diagnostic performance was observed across all viewpoints: area under the curves of the ROC to classify >=mild, >=moderate, =severe steatosis grades were 0.85, 0.90, and 0.93, respectively. The DL algorithm outperformed or performed at least comparably to FibroScan with statistically significant improvements for all levels on the unblinded histology-proven cohort, and for =severe steatosis on the blinded histology-proven cohort. Conclusions: The DL algorithm provides a reliable quantitative steatosis assessment across view and scanners on two multi-scanner cohorts. Diagnostic performance was high with comparable or better performance than FibroScan.
Learning from Subjective Ratings Using Auto-Decoded Deep Latent Embeddings
Apr 16, 2021



Abstract:Depending on the application, radiological diagnoses can be associated with high inter- and intra-rater variabilities. Most computer-aided diagnosis (CAD) solutions treat such data as incontrovertible, exposing learning algorithms to considerable and possibly contradictory label noise and biases. Thus, managing subjectivity in labels is a fundamental problem in medical imaging analysis. To address this challenge, we introduce auto-decoded deep latent embeddings (ADDLE), which explicitly models the tendencies of each rater using an auto-decoder framework. After a simple linear transformation, the latent variables can be injected into any backbone at any and multiple points, allowing the model to account for rater-specific effects on the diagnosis. Importantly, ADDLE does not expect multiple raters per image in training, meaning it can readily learn from data mined from hospital archives. Moreover, the complexity of training ADDLE does not increase as more raters are added. During inference each rater can be simulated and a 'mean' or 'greedy' virtual rating can be produced. We test ADDLE on the problem of liver steatosis diagnosis from 2D ultrasound (US) by collecting 46 084 studies along with clinical US diagnoses originating from 65 different raters. We evaluated diagnostic performance using a separate dataset with gold-standard biopsy diagnoses. ADDLE can improve the partial areas under the curve (AUCs) for diagnosing severe steatosis by 10.5% over standard classifiers while outperforming other annotator-noise approaches, including those requiring 65 times the parameters.
Reliable Liver Fibrosis Assessment from Ultrasound using Global Hetero-Image Fusion and View-Specific Parameterization
Aug 07, 2020



Abstract:Ultrasound (US) is a critical modality for diagnosing liver fibrosis. Unfortunately, assessment is very subjective, motivating automated approaches. We introduce a principled deep convolutional neural network (CNN) workflow that incorporates several innovations. First, to avoid overfitting on non-relevant image features, we force the network to focus on a clinical region of interest (ROI), encompassing the liver parenchyma and upper border. Second, we introduce global heteroimage fusion (GHIF), which allows the CNN to fuse features from any arbitrary number of images in a study, increasing its versatility and flexibility. Finally, we use 'style'-based view-specific parameterization (VSP) to tailor the CNN processing for different viewpoints of the liver, while keeping the majority of parameters the same across views. Experiments on a dataset of 610 patient studies (6979 images) demonstrate that our pipeline can contribute roughly 7% and 22% improvements in partial area under the curve and recall at 90% precision, respectively, over conventional classifiers, validating our approach to this crucial problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge