Jianping Lu
Streamlining Biomedical Research with Specialized LLMs
Apr 15, 2025Abstract:In this paper, we propose a novel system that integrates state-of-the-art, domain-specific large language models with advanced information retrieval techniques to deliver comprehensive and context-aware responses. Our approach facilitates seamless interaction among diverse components, enabling cross-validation of outputs to produce accurate, high-quality responses enriched with relevant data, images, tables, and other modalities. We demonstrate the system's capability to enhance response precision by leveraging a robust question-answering model, significantly improving the quality of dialogue generation. The system provides an accessible platform for real-time, high-fidelity interactions, allowing users to benefit from efficient human-computer interaction, precise retrieval, and simultaneous access to a wide range of literature and data. This dramatically improves the research efficiency of professionals in the biomedical and pharmaceutical domains and facilitates faster, more informed decision-making throughout the R\&D process. Furthermore, the system proposed in this paper is available at https://synapse-chat.patsnap.com.
PharmaGPT: Domain-Specific Large Language Models for Bio-Pharmaceutical and Chemistry
Jul 03, 2024



Abstract:Large language models (LLMs) have revolutionized Natural Language Processing (NLP) by by minimizing the need for complex feature engineering. However, the application of LLMs in specialized domains like biopharmaceuticals and chemistry remains largely unexplored. These fields are characterized by intricate terminologies, specialized knowledge, and a high demand for precision areas where general purpose LLMs often fall short. In this study, we introduce PharmGPT, a suite of multilingual LLMs with 13 billion and 70 billion parameters, specifically trained on a comprehensive corpus of hundreds of billions of tokens tailored to the Bio-Pharmaceutical and Chemical sectors. Our evaluation shows that PharmGPT matches or surpasses existing general models on key benchmarks, such as NAPLEX, demonstrating its exceptional capability in domain-specific tasks. This advancement establishes a new benchmark for LLMs in the Bio-Pharmaceutical and Chemical fields, addressing the existing gap in specialized language modeling. Furthermore, this suggests a promising path for enhanced research and development in these specialized areas, paving the way for more precise and effective applications of NLP in specialized domains.
PharmGPT: Domain-Specific Large Language Models for Bio-Pharmaceutical and Chemistry
Jun 26, 2024



Abstract:Large language models (LLMs) have revolutionized Natural Language Processing (NLP) by by minimizing the need for complex feature engineering. However, the application of LLMs in specialized domains like biopharmaceuticals and chemistry remains largely unexplored. These fields are characterized by intricate terminologies, specialized knowledge, and a high demand for precision areas where general purpose LLMs often fall short. In this study, we introduce PharmGPT, a suite of multilingual LLMs with 13 billion and 70 billion parameters, specifically trained on a comprehensive corpus of hundreds of billions of tokens tailored to the Bio-Pharmaceutical and Chemical sectors. Our evaluation shows that PharmGPT matches or surpasses existing general models on key benchmarks, such as NAPLEX, demonstrating its exceptional capability in domain-specific tasks. This advancement establishes a new benchmark for LLMs in the Bio-Pharmaceutical and Chemical fields, addressing the existing gap in specialized language modeling. Furthermore, this suggests a promising path for enhanced research and development in these specialized areas, paving the way for more precise and effective applications of NLP in specialized domains.
PatentGPT: A Large Language Model for Intellectual Property
Apr 30, 2024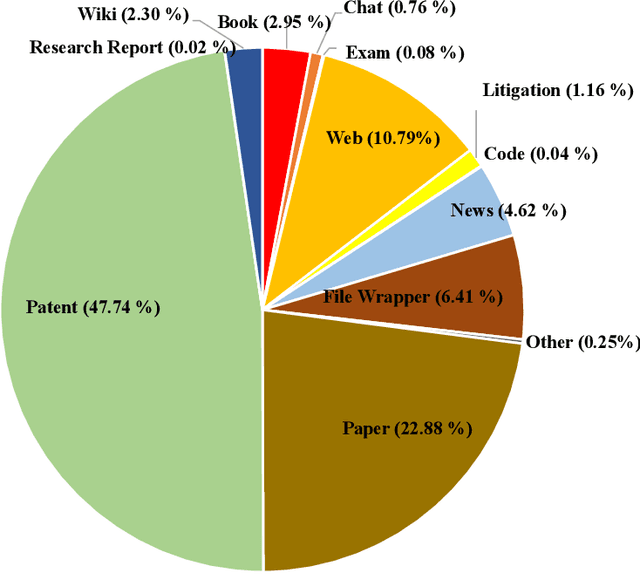

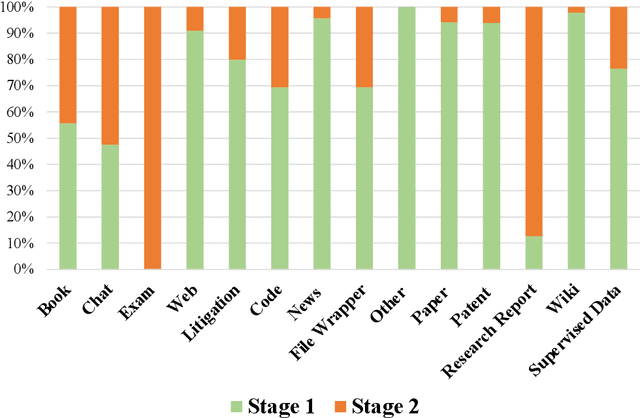
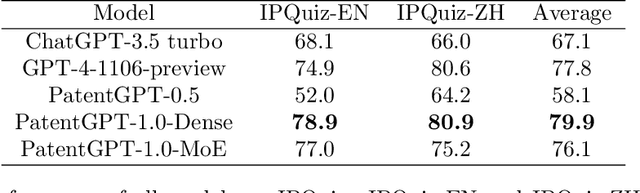
Abstract:In recent years, large language models have attracted significant attention due to their exceptional performance across a multitude of natural language process tasks, and have been widely applied in various fields. However, the application of large language models in the Intellectual Property (IP) space is challenging due to the strong need for specialized knowledge, privacy protection, processing of extremely long text in this field. In this technical report, we present for the first time a low-cost, standardized procedure for training IP-oriented LLMs, meeting the unique requirements of the IP domain. Using this standard process, we have trained the PatentGPT series models based on open-source pretrained models. By evaluating them on the open-source IP-oriented benchmark MOZIP, our domain-specific LLMs outperforms GPT-4, indicating the effectiveness of the proposed training procedure and the expertise of the PatentGPT models in the IP demain. What is impressive is that our model significantly outperformed GPT-4 on the 2019 China Patent Agent Qualification Examination by achieving a score of 65, reaching the level of human experts. Additionally, the PatentGPT model, which utilizes the SMoE architecture, achieves performance comparable to that of GPT-4 in the IP domain and demonstrates a better cost-performance ratio on long-text tasks, potentially serving as an alternative to GPT-4 within the IP domain.
A deep local attention network for pre-operative lymph node metastasis prediction in pancreatic cancer via multiphase CT imaging
Jan 04, 2023



Abstract:Lymph node (LN) metastasis status is one of the most critical prognostic and cancer staging factors for patients with resectable pancreatic ductal adenocarcinoma (PDAC), or in general, for any types of solid malignant tumors. Preoperative prediction of LN metastasis from non-invasive CT imaging is highly desired, as it might be straightforwardly used to guide the following neoadjuvant treatment decision and surgical planning. Most studies only capture the tumor characteristics in CT imaging to implicitly infer LN metastasis and very few work exploit direct LN's CT imaging information. To the best of our knowledge, this is the first work to propose a fully-automated LN segmentation and identification network to directly facilitate the LN metastasis status prediction task. Nevertheless LN segmentation/detection is very challenging since LN can be easily confused with other hard negative anatomic structures (e.g., vessels) from radiological images. We explore the anatomical spatial context priors of pancreatic LN locations by generating a guiding attention map from related organs and vessels to assist segmentation and infer LN status. As such, LN segmentation is impelled to focus on regions that are anatomically adjacent or plausible with respect to the specific organs and vessels. The metastasized LN identification network is trained to classify the segmented LN instances into positives or negatives by reusing the segmentation network as a pre-trained backbone and padding a new classification head. More importantly, we develop a LN metastasis status prediction network that combines the patient-wise aggregation results of LN segmentation/identification and deep imaging features extracted from the tumor region. Extensive quantitative nested five-fold cross-validation is conducted on a discovery dataset of 749 patients with PDAC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge