Yuan Zhong
A Computational Method for Solving the Stochastic Joint Replenishment Problem in High Dimensions
Nov 14, 2025Abstract:We consider a discrete-time formulation for a class of high-dimensional stochastic joint replenishment problems. First, we approximate the problem by a continuous-time impulse control problem. Exploiting connections among the impulse control problem, backward stochastic differential equations (BSDEs) with jumps, and the stochastic target problem, we develop a novel, simulation-based computational method that relies on deep neural networks to solve the impulse control problem. Based on that solution, we propose an implementable inventory control policy for the original (discrete-time) stochastic joint replenishment problem, and test it against the best available benchmarks in a series of test problems. For the problems studied thus far, our method matches or beats the best benchmark we could find, and it is computationally feasible up to at least 50 dimensions -- that is, 50 stock-keeping units (SKUs).
Two-Dimensional Pinching-Antenna Systems: Modeling and Beamforming Design
Nov 12, 2025



Abstract:Recently, the pinching-antenna system (PASS) has emerged as a promising architecture owing to its ability to reconfigure large-scale path loss and signal phase by activating radiation points along a dielectric waveguide. However, existing studies mainly focus on line-shaped PASS architectures, whose limited spatial flexibility constrains their applicability in multiuser and indoor scenarios. In this paper, we propose a novel two-dimensional (2D) pinching-antenna system (2D-PASS) that extends the conventional line-shaped structure into a continuous dielectric waveguide plane, thereby forming a reconfigurable radiating plane capable of dynamic beam adaptation across a 2D spatial domain. An optimization framework is developed to maximize the minimum received signal-to-noise ratio (SNR) among user equipments (UEs) by adaptively adjusting the spatial configuration of pinching antennas (PAs), serving as an analog beamforming mechanism for dynamic spatial control. For the continuous-position scenario, a particle swarm optimization (PSO)-based algorithm is proposed to efficiently explore the nonconvex search space, while a discrete variant is introduced to accommodate practical hardware constraints with limited PA placement resolution. Simulation results demonstrate that the proposed 2D-PASS substantially improves the minimum SNR compared with conventional line-shaped PASS and fixed-position antenna (FPA) benchmarks, while maintaining robustness under varying user distributions and distances.
MEDMKG: Benchmarking Medical Knowledge Exploitation with Multimodal Knowledge Graph
May 22, 2025Abstract:Medical deep learning models depend heavily on domain-specific knowledge to perform well on knowledge-intensive clinical tasks. Prior work has primarily leveraged unimodal knowledge graphs, such as the Unified Medical Language System (UMLS), to enhance model performance. However, integrating multimodal medical knowledge graphs remains largely underexplored, mainly due to the lack of resources linking imaging data with clinical concepts. To address this gap, we propose MEDMKG, a Medical Multimodal Knowledge Graph that unifies visual and textual medical information through a multi-stage construction pipeline. MEDMKG fuses the rich multimodal data from MIMIC-CXR with the structured clinical knowledge from UMLS, utilizing both rule-based tools and large language models for accurate concept extraction and relationship modeling. To ensure graph quality and compactness, we introduce Neighbor-aware Filtering (NaF), a novel filtering algorithm tailored for multimodal knowledge graphs. We evaluate MEDMKG across three tasks under two experimental settings, benchmarking twenty-four baseline methods and four state-of-the-art vision-language backbones on six datasets. Results show that MEDMKG not only improves performance in downstream medical tasks but also offers a strong foundation for developing adaptive and robust strategies for multimodal knowledge integration in medical artificial intelligence.
FairMedFM: Fairness Benchmarking for Medical Imaging Foundation Models
Jul 01, 2024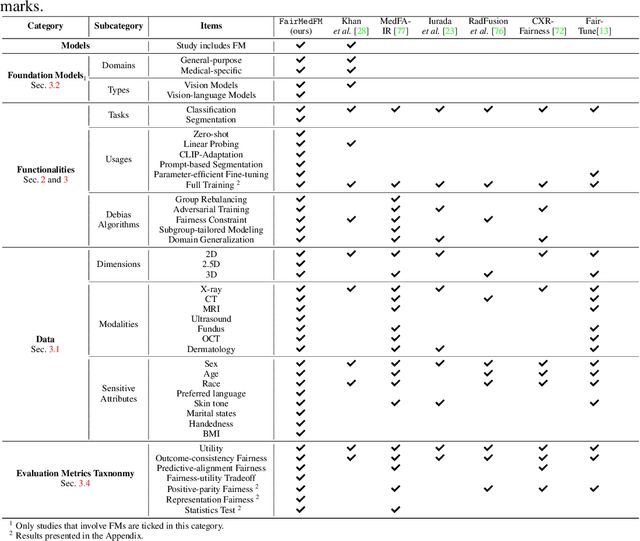
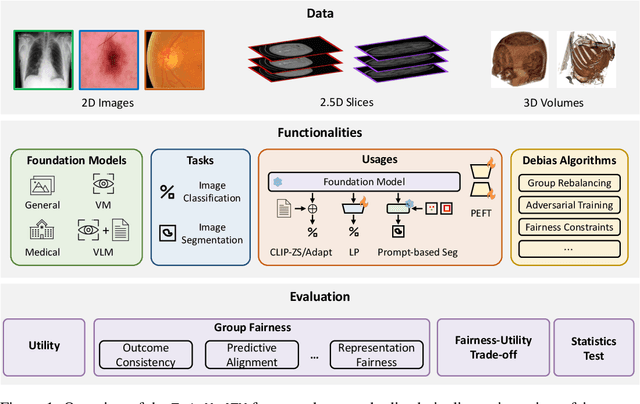
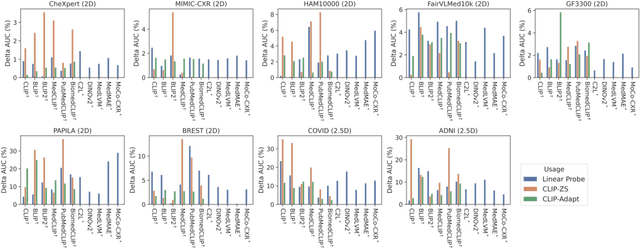
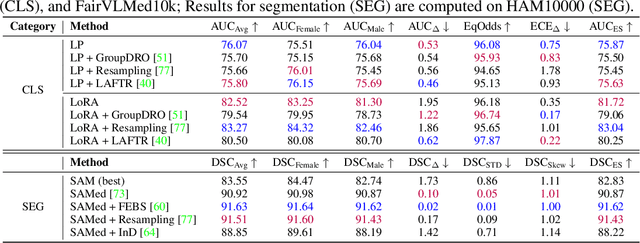
Abstract:The advent of foundation models (FMs) in healthcare offers unprecedented opportunities to enhance medical diagnostics through automated classification and segmentation tasks. However, these models also raise significant concerns about their fairness, especially when applied to diverse and underrepresented populations in healthcare applications. Currently, there is a lack of comprehensive benchmarks, standardized pipelines, and easily adaptable libraries to evaluate and understand the fairness performance of FMs in medical imaging, leading to considerable challenges in formulating and implementing solutions that ensure equitable outcomes across diverse patient populations. To fill this gap, we introduce FairMedFM, a fairness benchmark for FM research in medical imaging.FairMedFM integrates with 17 popular medical imaging datasets, encompassing different modalities, dimensionalities, and sensitive attributes. It explores 20 widely used FMs, with various usages such as zero-shot learning, linear probing, parameter-efficient fine-tuning, and prompting in various downstream tasks -- classification and segmentation. Our exhaustive analysis evaluates the fairness performance over different evaluation metrics from multiple perspectives, revealing the existence of bias, varied utility-fairness trade-offs on different FMs, consistent disparities on the same datasets regardless FMs, and limited effectiveness of existing unfairness mitigation methods. Checkout FairMedFM's project page and open-sourced codebase, which supports extendible functionalities and applications as well as inclusive for studies on FMs in medical imaging over the long term.
Synthesizing Multimodal Electronic Health Records via Predictive Diffusion Models
Jun 20, 2024



Abstract:Synthesizing electronic health records (EHR) data has become a preferred strategy to address data scarcity, improve data quality, and model fairness in healthcare. However, existing approaches for EHR data generation predominantly rely on state-of-the-art generative techniques like generative adversarial networks, variational autoencoders, and language models. These methods typically replicate input visits, resulting in inadequate modeling of temporal dependencies between visits and overlooking the generation of time information, a crucial element in EHR data. Moreover, their ability to learn visit representations is limited due to simple linear mapping functions, thus compromising generation quality. To address these limitations, we propose a novel EHR data generation model called EHRPD. It is a diffusion-based model designed to predict the next visit based on the current one while also incorporating time interval estimation. To enhance generation quality and diversity, we introduce a novel time-aware visit embedding module and a pioneering predictive denoising diffusion probabilistic model (PDDPM). Additionally, we devise a predictive U-Net (PU-Net) to optimize P-DDPM.We conduct experiments on two public datasets and evaluate EHRPD from fidelity, privacy, and utility perspectives. The experimental results demonstrate the efficacy and utility of the proposed EHRPD in addressing the aforementioned limitations and advancing EHR data generation.
PatentGPT: A Large Language Model for Intellectual Property
Apr 30, 2024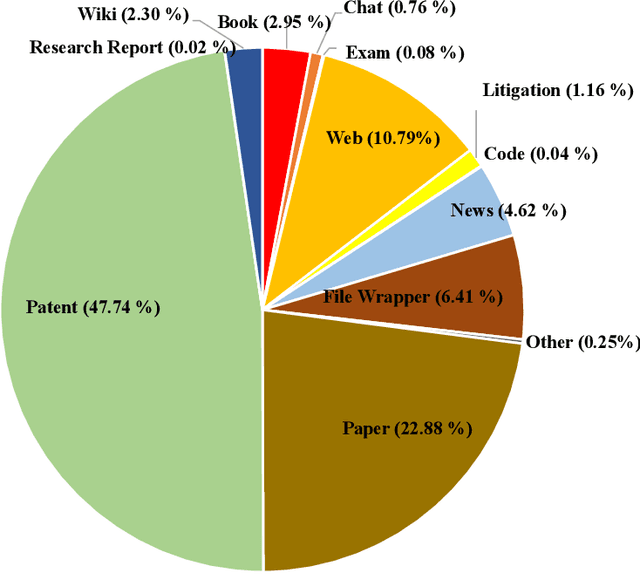

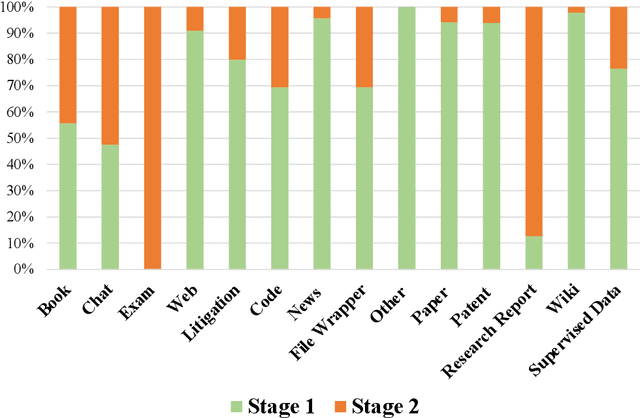
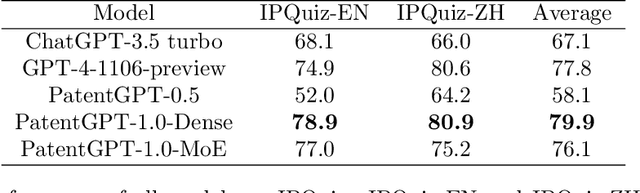
Abstract:In recent years, large language models have attracted significant attention due to their exceptional performance across a multitude of natural language process tasks, and have been widely applied in various fields. However, the application of large language models in the Intellectual Property (IP) space is challenging due to the strong need for specialized knowledge, privacy protection, processing of extremely long text in this field. In this technical report, we present for the first time a low-cost, standardized procedure for training IP-oriented LLMs, meeting the unique requirements of the IP domain. Using this standard process, we have trained the PatentGPT series models based on open-source pretrained models. By evaluating them on the open-source IP-oriented benchmark MOZIP, our domain-specific LLMs outperforms GPT-4, indicating the effectiveness of the proposed training procedure and the expertise of the PatentGPT models in the IP demain. What is impressive is that our model significantly outperformed GPT-4 on the 2019 China Patent Agent Qualification Examination by achieving a score of 65, reaching the level of human experts. Additionally, the PatentGPT model, which utilizes the SMoE architecture, achieves performance comparable to that of GPT-4 in the IP domain and demonstrates a better cost-performance ratio on long-text tasks, potentially serving as an alternative to GPT-4 within the IP domain.
FineRec:Exploring Fine-grained Sequential Recommendation
Apr 19, 2024



Abstract:Sequential recommendation is dedicated to offering items of interest for users based on their history behaviors. The attribute-opinion pairs, expressed by users in their reviews for items, provide the potentials to capture user preferences and item characteristics at a fine-grained level. To this end, we propose a novel framework FineRec that explores the attribute-opinion pairs of reviews to finely handle sequential recommendation. Specifically, we utilize a large language model to extract attribute-opinion pairs from reviews. For each attribute, a unique attribute-specific user-opinion-item graph is created, where corresponding opinions serve as the edges linking heterogeneous user and item nodes. To tackle the diversity of opinions, we devise a diversity-aware convolution operation to aggregate information within the graphs, enabling attribute-specific user and item representation learning. Ultimately, we present an interaction-driven fusion mechanism to integrate attribute-specific user/item representations across all attributes for generating recommendations. Extensive experiments conducted on several realworld datasets demonstrate the superiority of our FineRec over existing state-of-the-art methods. Further analysis also verifies the effectiveness of our fine-grained manner in handling the task.
Recent Advances in Predictive Modeling with Electronic Health Records
Feb 02, 2024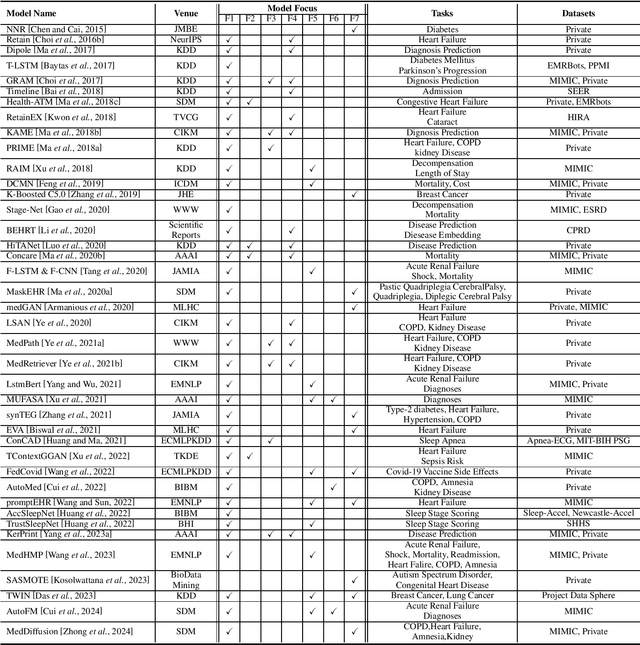
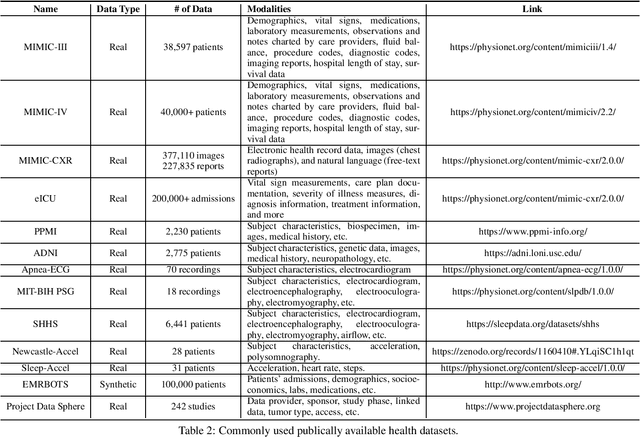
Abstract:The development of electronic health records (EHR) systems has enabled the collection of a vast amount of digitized patient data. However, utilizing EHR data for predictive modeling presents several challenges due to its unique characteristics. With the advancements in machine learning techniques, deep learning has demonstrated its superiority in various applications, including healthcare. This survey systematically reviews recent advances in deep learning-based predictive models using EHR data. Specifically, we begin by introducing the background of EHR data and providing a mathematical definition of the predictive modeling task. We then categorize and summarize predictive deep models from multiple perspectives. Furthermore, we present benchmarks and toolkits relevant to predictive modeling in healthcare. Finally, we conclude this survey by discussing open challenges and suggesting promising directions for future research.
Automated Fusion of Multimodal Electronic Health Records for Better Medical Predictions
Jan 20, 2024Abstract:The widespread adoption of Electronic Health Record (EHR) systems in healthcare institutes has generated vast amounts of medical data, offering significant opportunities for improving healthcare services through deep learning techniques. However, the complex and diverse modalities and feature structures in real-world EHR data pose great challenges for deep learning model design. To address the multi-modality challenge in EHR data, current approaches primarily rely on hand-crafted model architectures based on intuition and empirical experiences, leading to sub-optimal model architectures and limited performance. Therefore, to automate the process of model design for mining EHR data, we propose a novel neural architecture search (NAS) framework named AutoFM, which can automatically search for the optimal model architectures for encoding diverse input modalities and fusion strategies. We conduct thorough experiments on real-world multi-modal EHR data and prediction tasks, and the results demonstrate that our framework not only achieves significant performance improvement over existing state-of-the-art methods but also discovers meaningful network architectures effectively.
Hierarchical Pretraining on Multimodal Electronic Health Records
Oct 20, 2023Abstract:Pretraining has proven to be a powerful technique in natural language processing (NLP), exhibiting remarkable success in various NLP downstream tasks. However, in the medical domain, existing pretrained models on electronic health records (EHR) fail to capture the hierarchical nature of EHR data, limiting their generalization capability across diverse downstream tasks using a single pretrained model. To tackle this challenge, this paper introduces a novel, general, and unified pretraining framework called MEDHMP, specifically designed for hierarchically multimodal EHR data. The effectiveness of the proposed MEDHMP is demonstrated through experimental results on eight downstream tasks spanning three levels. Comparisons against eighteen baselines further highlight the efficacy of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge