Yinda Chen
MAIN-VLA: Modeling Abstraction of Intention and eNvironment for Vision-Language-Action Models
Feb 02, 2026Abstract:Despite significant progress in Visual-Language-Action (VLA), in highly complex and dynamic environments that involve real-time unpredictable interactions (such as 3D open worlds and large-scale PvP games), existing approaches remain inefficient at extracting action-critical signals from redundant sensor streams. To tackle this, we introduce MAIN-VLA, a framework that explicitly Models the Abstraction of Intention and eNvironment to ground decision-making in deep semantic alignment rather than superficial pattern matching. Specifically, our Intention Abstraction (IA) extracts verbose linguistic instructions and their associated reasoning into compact, explicit semantic primitives, while the Environment Semantics Abstraction (ESA) projects overwhelming visual streams into a structured, topological affordance representation. Furthermore, aligning these two abstract modalities induces an emergent attention-concentration effect, enabling a parameter-free token-pruning strategy that filters out perceptual redundancy without degrading performance. Extensive experiments in open-world Minecraft and large-scale PvP environments (Game for Peace and Valorant) demonstrate that MAIN-VLA sets a new state-of-the-art, which achieves superior decision quality, stronger generalization, and cutting-edge inference efficiency.
Johnson-Lindenstrauss Lemma Guided Network for Efficient 3D Medical Segmentation
Sep 26, 2025Abstract:Lightweight 3D medical image segmentation remains constrained by a fundamental "efficiency / robustness conflict", particularly when processing complex anatomical structures and heterogeneous modalities. In this paper, we study how to redesign the framework based on the characteristics of high-dimensional 3D images, and explore data synergy to overcome the fragile representation of lightweight methods. Our approach, VeloxSeg, begins with a deployable and extensible dual-stream CNN-Transformer architecture composed of Paired Window Attention (PWA) and Johnson-Lindenstrauss lemma-guided convolution (JLC). For each 3D image, we invoke a "glance-and-focus" principle, where PWA rapidly retrieves multi-scale information, and JLC ensures robust local feature extraction with minimal parameters, significantly enhancing the model's ability to operate with low computational budget. Followed by an extension of the dual-stream architecture that incorporates modal interaction into the multi-scale image-retrieval process, VeloxSeg efficiently models heterogeneous modalities. Finally, Spatially Decoupled Knowledge Transfer (SDKT) via Gram matrices injects the texture prior extracted by a self-supervised network into the segmentation network, yielding stronger representations than baselines at no extra inference cost. Experimental results on multimodal benchmarks show that VeloxSeg achieves a 26% Dice improvement, alongside increasing GPU throughput by 11x and CPU by 48x. Codes are available at https://github.com/JinPLu/VeloxSeg.
FURINA: Free from Unmergeable Router via LINear Aggregation of mixed experts
Sep 18, 2025Abstract:The Mixture of Experts (MoE) paradigm has been successfully integrated into Low-Rank Adaptation (LoRA) for parameter-efficient fine-tuning (PEFT), delivering performance gains with minimal parameter overhead. However, a key limitation of existing MoE-LoRA methods is their reliance on a discrete router, which prevents the integration of the MoE components into the backbone model. To overcome this, we propose FURINA, a novel Free from Unmergeable Router framework based on the LINear Aggregation of experts. FURINA eliminates the router by introducing a Self-Routing mechanism. This is achieved through three core innovations: (1) decoupled learning of the direction and magnitude for LoRA adapters, (2) a shared learnable magnitude vector for consistent activation scaling, and (3) expert selection loss that encourages divergent expert activation. The proposed mechanism leverages the angular similarity between the input and each adapter's directional component to activate experts, which are then scaled by the shared magnitude vector. This design allows the output norm to naturally reflect the importance of each expert, thereby enabling dynamic, router-free routing. The expert selection loss further sharpens this behavior by encouraging sparsity and aligning it with standard MoE activation patterns. We also introduce a shared expert within the MoE-LoRA block that provides stable, foundational knowledge. To the best of our knowledge, FURINA is the first router-free, MoE-enhanced LoRA method that can be fully merged into the backbone model, introducing zero additional inference-time cost or complexity. Extensive experiments demonstrate that FURINA not only significantly outperforms standard LoRA but also matches or surpasses the performance of existing MoE-LoRA methods, while eliminating the extra inference-time overhead of MoE.
EMPOWER: Evolutionary Medical Prompt Optimization With Reinforcement Learning
Aug 25, 2025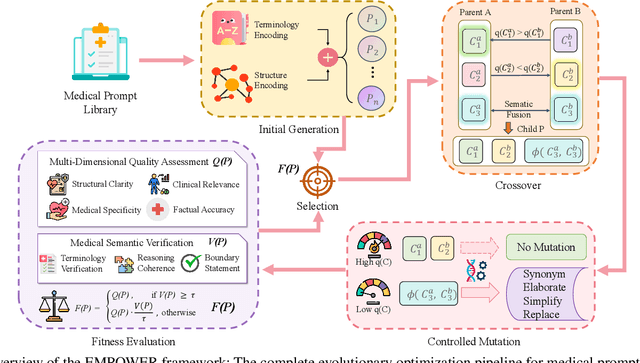
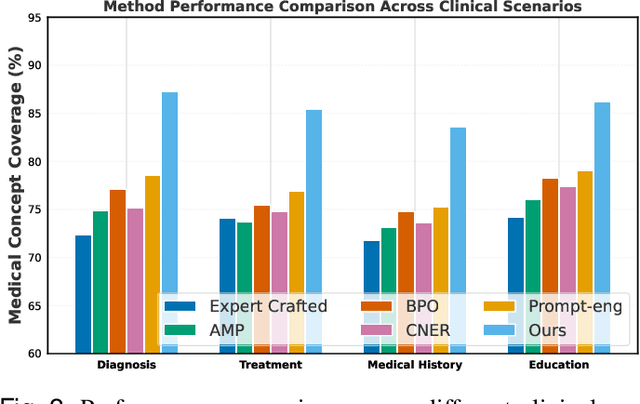
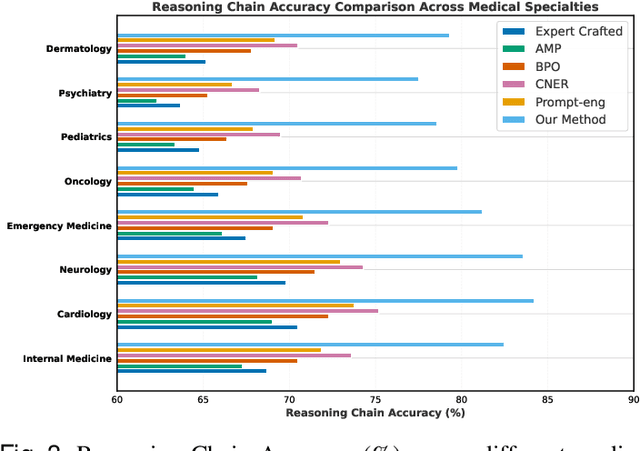
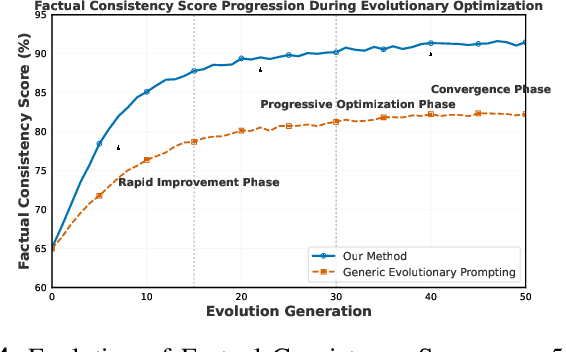
Abstract:Prompt engineering significantly influences the reliability and clinical utility of Large Language Models (LLMs) in medical applications. Current optimization approaches inadequately address domain-specific medical knowledge and safety requirements. This paper introduces EMPOWER, a novel evolutionary framework that enhances medical prompt quality through specialized representation learning, multi-dimensional evaluation, and structure-preserving algorithms. Our methodology incorporates: (1) a medical terminology attention mechanism, (2) a comprehensive assessment architecture evaluating clarity, specificity, clinical relevance, and factual accuracy, (3) a component-level evolutionary algorithm preserving clinical reasoning integrity, and (4) a semantic verification module ensuring adherence to medical knowledge. Evaluation across diagnostic, therapeutic, and educational tasks demonstrates significant improvements: 24.7% reduction in factually incorrect content, 19.6% enhancement in domain specificity, and 15.3% higher clinician preference in blinded evaluations. The framework addresses critical challenges in developing clinically appropriate prompts, facilitating more responsible integration of LLMs into healthcare settings.
Low-Cost Test-Time Adaptation for Robust Video Editing
Jul 29, 2025



Abstract:Video editing is a critical component of content creation that transforms raw footage into coherent works aligned with specific visual and narrative objectives. Existing approaches face two major challenges: temporal inconsistencies due to failure in capturing complex motion patterns, and overfitting to simple prompts arising from limitations in UNet backbone architectures. While learning-based methods can enhance editing quality, they typically demand substantial computational resources and are constrained by the scarcity of high-quality annotated data. In this paper, we present Vid-TTA, a lightweight test-time adaptation framework that personalizes optimization for each test video during inference through self-supervised auxiliary tasks. Our approach incorporates a motion-aware frame reconstruction mechanism that identifies and preserves crucial movement regions, alongside a prompt perturbation and reconstruction strategy that strengthens model robustness to diverse textual descriptions. These innovations are orchestrated by a meta-learning driven dynamic loss balancing mechanism that adaptively adjusts the optimization process based on video characteristics. Extensive experiments demonstrate that Vid-TTA significantly improves video temporal consistency and mitigates prompt overfitting while maintaining low computational overhead, offering a plug-and-play performance boost for existing video editing models.
Dual form Complementary Masking for Domain-Adaptive Image Segmentation
Jul 16, 2025Abstract:Recent works have correlated Masked Image Modeling (MIM) with consistency regularization in Unsupervised Domain Adaptation (UDA). However, they merely treat masking as a special form of deformation on the input images and neglect the theoretical analysis, which leads to a superficial understanding of masked reconstruction and insufficient exploitation of its potential in enhancing feature extraction and representation learning. In this paper, we reframe masked reconstruction as a sparse signal reconstruction problem and theoretically prove that the dual form of complementary masks possesses superior capabilities in extracting domain-agnostic image features. Based on this compelling insight, we propose MaskTwins, a simple yet effective UDA framework that integrates masked reconstruction directly into the main training pipeline. MaskTwins uncovers intrinsic structural patterns that persist across disparate domains by enforcing consistency between predictions of images masked in complementary ways, enabling domain generalization in an end-to-end manner. Extensive experiments verify the superiority of MaskTwins over baseline methods in natural and biological image segmentation. These results demonstrate the significant advantages of MaskTwins in extracting domain-invariant features without the need for separate pre-training, offering a new paradigm for domain-adaptive segmentation.
DVP-MVS++: Synergize Depth-Normal-Edge and Harmonized Visibility Prior for Multi-View Stereo
Jun 16, 2025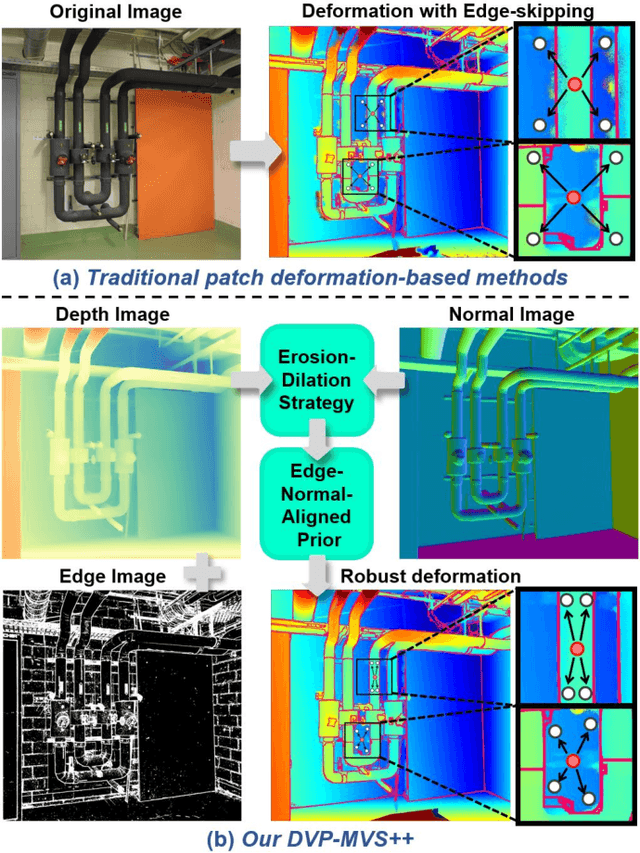
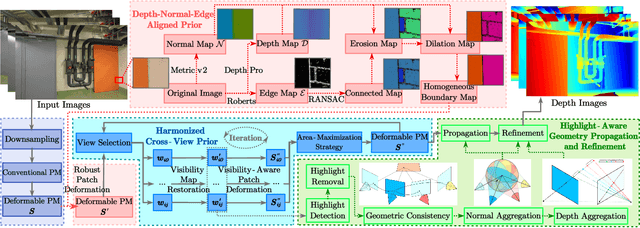
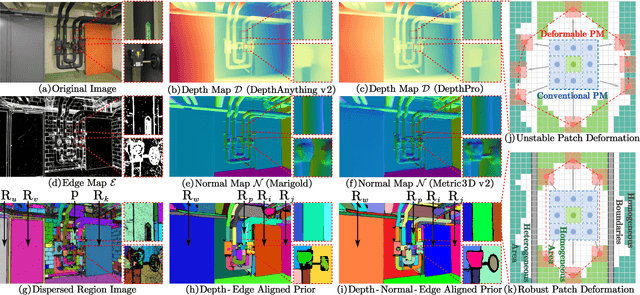
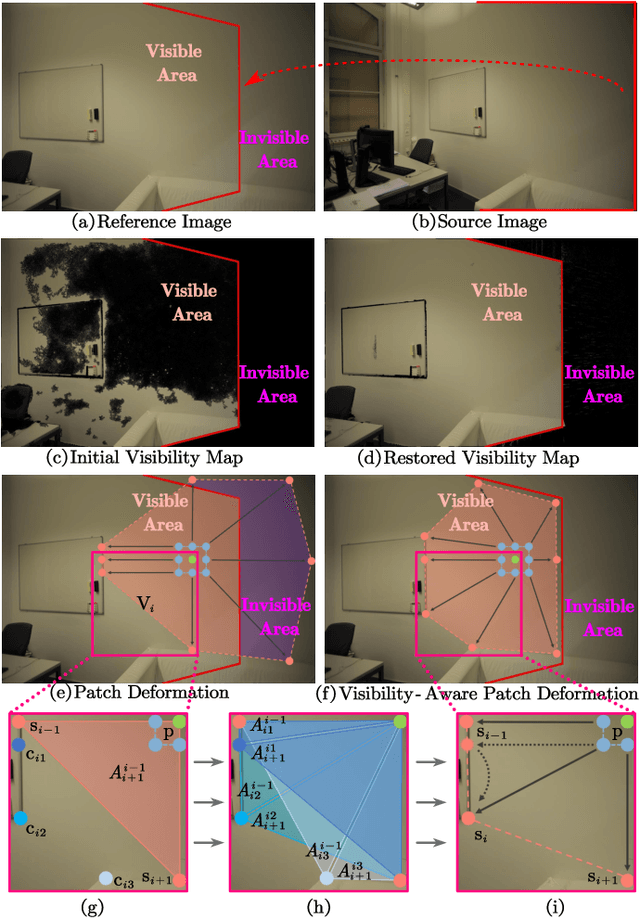
Abstract:Recently, patch deformation-based methods have demonstrated significant effectiveness in multi-view stereo due to their incorporation of deformable and expandable perception for reconstructing textureless areas. However, these methods generally focus on identifying reliable pixel correlations to mitigate matching ambiguity of patch deformation, while neglecting the deformation instability caused by edge-skipping and visibility occlusions, which may cause potential estimation deviations. To address these issues, we propose DVP-MVS++, an innovative approach that synergizes both depth-normal-edge aligned and harmonized cross-view priors for robust and visibility-aware patch deformation. Specifically, to avoid edge-skipping, we first apply DepthPro, Metric3Dv2 and Roberts operator to generate coarse depth maps, normal maps and edge maps, respectively. These maps are then aligned via an erosion-dilation strategy to produce fine-grained homogeneous boundaries for facilitating robust patch deformation. Moreover, we reformulate view selection weights as visibility maps, and then implement both an enhanced cross-view depth reprojection and an area-maximization strategy to help reliably restore visible areas and effectively balance deformed patch, thus acquiring harmonized cross-view priors for visibility-aware patch deformation. Additionally, we obtain geometry consistency by adopting both aggregated normals via view selection and projection depth differences via epipolar lines, and then employ SHIQ for highlight correction to enable geometry consistency with highlight-aware perception, thus improving reconstruction quality during propagation and refinement stage. Evaluation results on ETH3D, Tanks & Temples and Strecha datasets exhibit the state-of-the-art performance and robust generalization capability of our proposed method.
QMamba: Post-Training Quantization for Vision State Space Models
Jan 23, 2025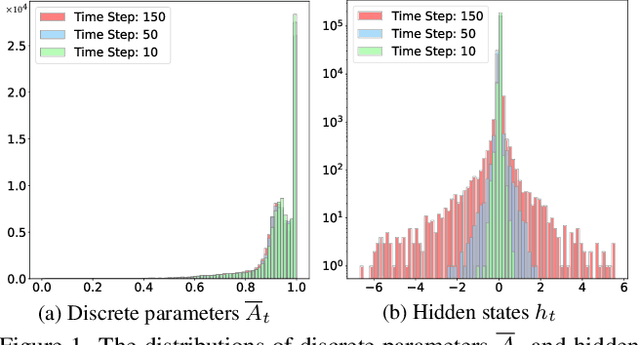

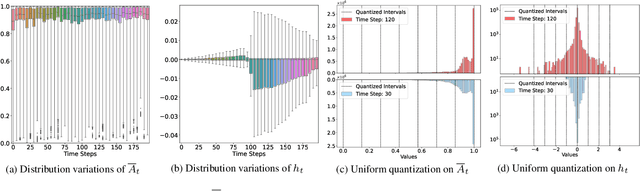

Abstract:State Space Models (SSMs), as key components of Mamaba, have gained increasing attention for vision models recently, thanks to their efficient long sequence modeling capability. Given the computational cost of deploying SSMs on resource-limited edge devices, Post-Training Quantization (PTQ) is a technique with the potential for efficient deployment of SSMs. In this work, we propose QMamba, one of the first PTQ frameworks to our knowledge, designed for vision SSMs based on the analysis of the activation distributions in SSMs. We reveal that the distribution of discrete parameters exhibits long-tailed skewness and the distribution of the hidden state sequence exhibits highly dynamic variations. Correspondingly, we design Long-tailed Skewness Quantization (LtSQ) to quantize discrete parameters and Temporal Group Quantization (TGQ) to quantize hidden states, which reduces the quantization errors. Extensive experiments demonstrate that QMamba outperforms advanced PTQ methods on vision models across multiple model sizes and architectures. Notably, QMamba surpasses existing methods by 21.0% on ImageNet classification with 4-bit activations.
MapExpert: Online HD Map Construction with Simple and Efficient Sparse Map Element Expert
Dec 17, 2024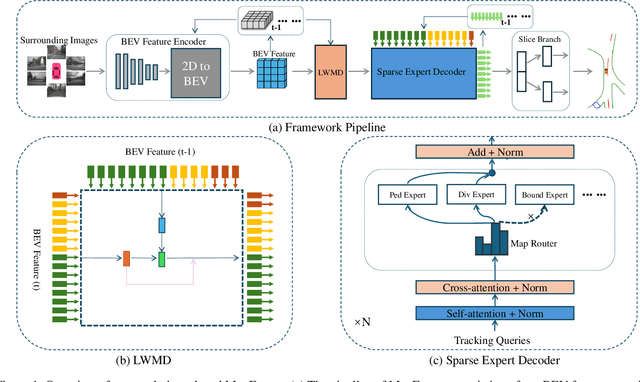
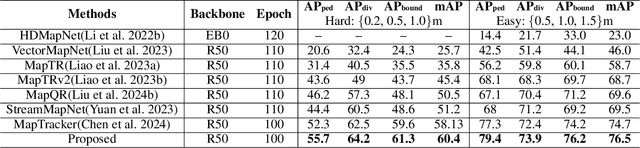

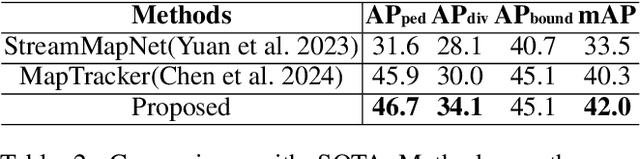
Abstract:Constructing online High-Definition (HD) maps is crucial for the static environment perception of autonomous driving systems (ADS). Existing solutions typically attempt to detect vectorized HD map elements with unified models; however, these methods often overlook the distinct characteristics of different non-cubic map elements, making accurate distinction challenging. To address these issues, we introduce an expert-based online HD map method, termed MapExpert. MapExpert utilizes sparse experts, distributed by our routers, to describe various non-cubic map elements accurately. Additionally, we propose an auxiliary balance loss function to distribute the load evenly across experts. Furthermore, we theoretically analyze the limitations of prevalent bird's-eye view (BEV) feature temporal fusion methods and introduce an efficient temporal fusion module called Learnable Weighted Moving Descentage. This module effectively integrates relevant historical information into the final BEV features. Combined with an enhanced slice head branch, the proposed MapExpert achieves state-of-the-art performance and maintains good efficiency on both nuScenes and Argoverse2 datasets.
Can Medical Vision-Language Pre-training Succeed with Purely Synthetic Data?
Oct 17, 2024



Abstract:Medical Vision-Language Pre-training (MedVLP) has made significant progress in enabling zero-shot tasks for medical image understanding. However, training MedVLP models typically requires large-scale datasets with paired, high-quality image-text data, which are scarce in the medical domain. Recent advancements in Large Language Models (LLMs) and diffusion models have made it possible to generate large-scale synthetic image-text pairs. This raises the question: *Can MedVLP succeed using purely synthetic data?* To address this, we use off-the-shelf generative models to create synthetic radiology reports and paired Chest X-ray (CXR) images, and propose an automated pipeline to build a diverse, high-quality synthetic dataset, enabling a rigorous study that isolates model and training settings, focusing entirely from the data perspective. Our results show that MedVLP models trained *exclusively on synthetic data* outperform those trained on real data by **3.8%** in averaged AUC on zero-shot classification. Moreover, using a combination of synthetic and real data leads to a further improvement of **9.07%**. Additionally, MedVLP models trained on synthetic or mixed data consistently outperform those trained on real data in zero-shot grounding, as well as in fine-tuned classification and segmentation tasks. Our analysis suggests MedVLP trained on well-designed synthetic data can outperform models trained on real datasets, which may be limited by low-quality samples and long-tailed distributions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge