Kunlun He
Generalized Deep Multi-view Clustering via Causal Learning with Partially Aligned Cross-view Correspondence
Sep 19, 2025Abstract:Multi-view clustering (MVC) aims to explore the common clustering structure across multiple views. Many existing MVC methods heavily rely on the assumption of view consistency, where alignments for corresponding samples across different views are ordered in advance. However, real-world scenarios often present a challenge as only partial data is consistently aligned across different views, restricting the overall clustering performance. In this work, we consider the model performance decreasing phenomenon caused by data order shift (i.e., from fully to partially aligned) as a generalized multi-view clustering problem. To tackle this problem, we design a causal multi-view clustering network, termed CauMVC. We adopt a causal modeling approach to understand multi-view clustering procedure. To be specific, we formulate the partially aligned data as an intervention and multi-view clustering with partially aligned data as an post-intervention inference. However, obtaining invariant features directly can be challenging. Thus, we design a Variational Auto-Encoder for causal learning by incorporating an encoder from existing information to estimate the invariant features. Moreover, a decoder is designed to perform the post-intervention inference. Lastly, we design a contrastive regularizer to capture sample correlations. To the best of our knowledge, this paper is the first work to deal generalized multi-view clustering via causal learning. Empirical experiments on both fully and partially aligned data illustrate the strong generalization and effectiveness of CauMVC.
Towards Pre-trained Graph Condensation via Optimal Transport
Sep 18, 2025Abstract:Graph condensation (GC) aims to distill the original graph into a small-scale graph, mitigating redundancy and accelerating GNN training. However, conventional GC approaches heavily rely on rigid GNNs and task-specific supervision. Such a dependency severely restricts their reusability and generalization across various tasks and architectures. In this work, we revisit the goal of ideal GC from the perspective of GNN optimization consistency, and then a generalized GC optimization objective is derived, by which those traditional GC methods can be viewed nicely as special cases of this optimization paradigm. Based on this, Pre-trained Graph Condensation (PreGC) via optimal transport is proposed to transcend the limitations of task- and architecture-dependent GC methods. Specifically, a hybrid-interval graph diffusion augmentation is presented to suppress the weak generalization ability of the condensed graph on particular architectures by enhancing the uncertainty of node states. Meanwhile, the matching between optimal graph transport plan and representation transport plan is tactfully established to maintain semantic consistencies across source graph and condensed graph spaces, thereby freeing graph condensation from task dependencies. To further facilitate the adaptation of condensed graphs to various downstream tasks, a traceable semantic harmonizer from source nodes to condensed nodes is proposed to bridge semantic associations through the optimized representation transport plan in pre-training. Extensive experiments verify the superiority and versatility of PreGC, demonstrating its task-independent nature and seamless compatibility with arbitrary GNNs.
Intra-view and Inter-view Correlation Guided Multi-view Novel Class Discovery
Jul 16, 2025Abstract:In this paper, we address the problem of novel class discovery (NCD), which aims to cluster novel classes by leveraging knowledge from disjoint known classes. While recent advances have made significant progress in this area, existing NCD methods face two major limitations. First, they primarily focus on single-view data (e.g., images), overlooking the increasingly common multi-view data, such as multi-omics datasets used in disease diagnosis. Second, their reliance on pseudo-labels to supervise novel class clustering often results in unstable performance, as pseudo-label quality is highly sensitive to factors such as data noise and feature dimensionality. To address these challenges, we propose a novel framework named Intra-view and Inter-view Correlation Guided Multi-view Novel Class Discovery (IICMVNCD), which is the first attempt to explore NCD in multi-view setting so far. Specifically, at the intra-view level, leveraging the distributional similarity between known and novel classes, we employ matrix factorization to decompose features into view-specific shared base matrices and factor matrices. The base matrices capture distributional consistency among the two datasets, while the factor matrices model pairwise relationships between samples. At the inter-view level, we utilize view relationships among known classes to guide the clustering of novel classes. This includes generating predicted labels through the weighted fusion of factor matrices and dynamically adjusting view weights of known classes based on the supervision loss, which are then transferred to novel class learning. Experimental results validate the effectiveness of our proposed approach.
Angio-Diff: Learning a Self-Supervised Adversarial Diffusion Model for Angiographic Geometry Generation
Jun 24, 2025Abstract:Vascular diseases pose a significant threat to human health, with X-ray angiography established as the gold standard for diagnosis, allowing for detailed observation of blood vessels. However, angiographic X-rays expose personnel and patients to higher radiation levels than non-angiographic X-rays, which are unwanted. Thus, modality translation from non-angiographic to angiographic X-rays is desirable. Data-driven deep approaches are hindered by the lack of paired large-scale X-ray angiography datasets. While making high-quality vascular angiography synthesis crucial, it remains challenging. We find that current medical image synthesis primarily operates at pixel level and struggles to adapt to the complex geometric structure of blood vessels, resulting in unsatisfactory quality of blood vessel image synthesis, such as disconnections or unnatural curvatures. To overcome this issue, we propose a self-supervised method via diffusion models to transform non-angiographic X-rays into angiographic X-rays, mitigating data shortages for data-driven approaches. Our model comprises a diffusion model that learns the distribution of vascular data from diffusion latent, a generator for vessel synthesis, and a mask-based adversarial module. To enhance geometric accuracy, we propose a parametric vascular model to fit the shape and distribution of blood vessels. The proposed method contributes a pipeline and a synthetic dataset for X-ray angiography. We conducted extensive comparative and ablation experiments to evaluate the Angio-Diff. The results demonstrate that our method achieves state-of-the-art performance in synthetic angiography image quality and more accurately synthesizes the geometric structure of blood vessels. The code is available at https://github.com/zfw-cv/AngioDiff.
Dynamic Graph Condensation
Jun 16, 2025Abstract:Recent research on deep graph learning has shifted from static to dynamic graphs, motivated by the evolving behaviors observed in complex real-world systems. However, the temporal extension in dynamic graphs poses significant data efficiency challenges, including increased data volume, high spatiotemporal redundancy, and reliance on costly dynamic graph neural networks (DGNNs). To alleviate the concerns, we pioneer the study of dynamic graph condensation (DGC), which aims to substantially reduce the scale of dynamic graphs for data-efficient DGNN training. Accordingly, we propose DyGC, a novel framework that condenses the real dynamic graph into a compact version while faithfully preserving the inherent spatiotemporal characteristics. Specifically, to endow synthetic graphs with realistic evolving structures, a novel spiking structure generation mechanism is introduced. It draws on the dynamic behavior of spiking neurons to model temporally-aware connectivity in dynamic graphs. Given the tightly coupled spatiotemporal dependencies, DyGC proposes a tailored distribution matching approach that first constructs a semantically rich state evolving field for dynamic graphs, and then performs fine-grained spatiotemporal state alignment to guide the optimization of the condensed graph. Experiments across multiple dynamic graph datasets and representative DGNN architectures demonstrate the effectiveness of DyGC. Notably, our method retains up to 96.2% DGNN performance with only 0.5% of the original graph size, and achieves up to 1846 times training speedup.
Causally-informed Deep Learning towards Explainable and Generalizable Outcomes Prediction in Critical Care
Feb 04, 2025



Abstract:Recent advances in deep learning (DL) have prompted the development of high-performing early warning score (EWS) systems, predicting clinical deteriorations such as acute kidney injury, acute myocardial infarction, or circulatory failure. DL models have proven to be powerful tools for various tasks but come with the cost of lacking interpretability and limited generalizability, hindering their clinical applications. To develop a practical EWS system applicable to various outcomes, we propose causally-informed explainable early prediction model, which leverages causal discovery to identify the underlying causal relationships of prediction and thus owns two unique advantages: demonstrating the explicit interpretation of the prediction while exhibiting decent performance when applied to unfamiliar environments. Benefiting from these features, our approach achieves superior accuracy for 6 different critical deteriorations and achieves better generalizability across different patient groups, compared to various baseline algorithms. Besides, we provide explicit causal pathways to serve as references for assistant clinical diagnosis and potential interventions. The proposed approach enhances the practical application of deep learning in various medical scenarios.
A Joint Representation Using Continuous and Discrete Features for Cardiovascular Diseases Risk Prediction on Chest CT Scans
Oct 24, 2024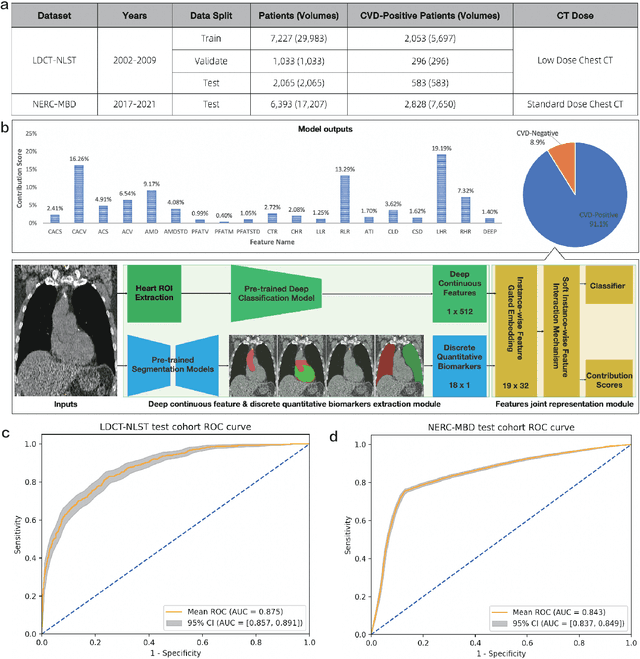
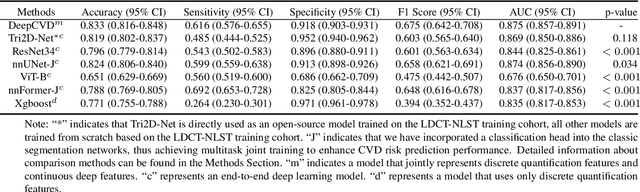
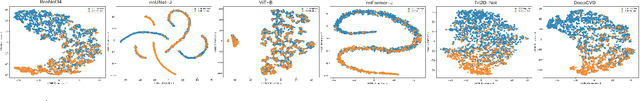
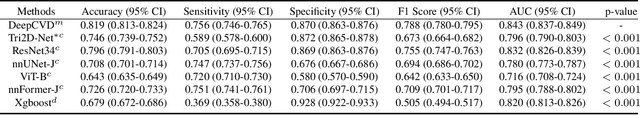
Abstract:Cardiovascular diseases (CVD) remain a leading health concern and contribute significantly to global mortality rates. While clinical advancements have led to a decline in CVD mortality, accurately identifying individuals who could benefit from preventive interventions remains an unsolved challenge in preventive cardiology. Current CVD risk prediction models, recommended by guidelines, are based on limited traditional risk factors or use CT imaging to acquire quantitative biomarkers, and still have limitations in predictive accuracy and applicability. On the other hand, end-to-end trained CVD risk prediction methods leveraging deep learning on CT images often fail to provide transparent and explainable decision grounds for assisting physicians. In this work, we proposed a novel joint representation that integrates discrete quantitative biomarkers and continuous deep features extracted from chest CT scans. Our approach initiated with a deep CVD risk classification model by capturing comprehensive continuous deep learning features while jointly obtaining currently clinical-established quantitative biomarkers via segmentation models. In the feature joint representation stage, we use an instance-wise feature-gated mechanism to align the continuous and discrete features, followed by a soft instance-wise feature interaction mechanism fostering independent and effective feature interaction for the final CVD risk prediction. Our method substantially improves CVD risk predictive performance and offers individual contribution analysis of each biomarker, which is important in assisting physicians' decision-making processes. We validated our method on a public chest low-dose CT dataset and a private external chest standard-dose CT patient cohort of 17,207 CT volumes from 6,393 unique subjects, and demonstrated superior predictive performance, achieving AUCs of 0.875 and 0.843, respectively.
Active-Passive Federated Learning for Vertically Partitioned Multi-view Data
Sep 06, 2024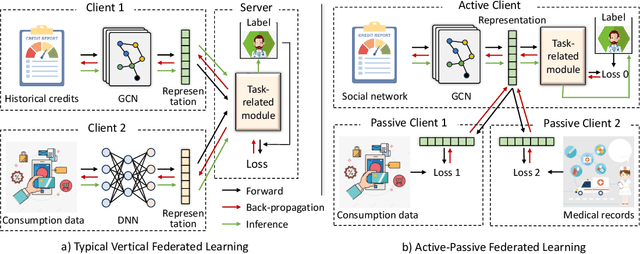


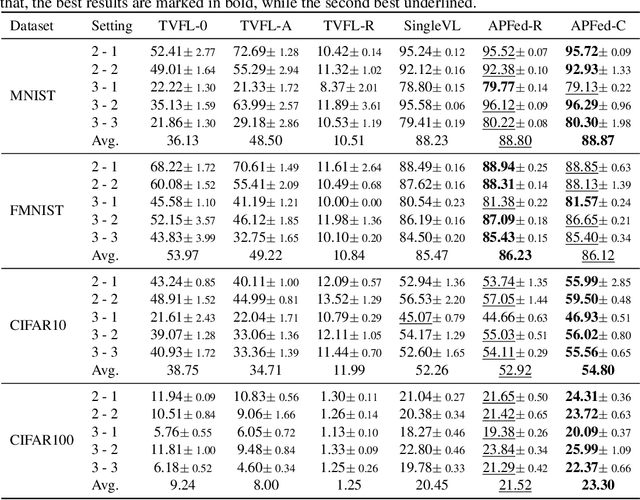
Abstract:Vertical federated learning is a natural and elegant approach to integrate multi-view data vertically partitioned across devices (clients) while preserving their privacies. Apart from the model training, existing methods requires the collaboration of all clients in the model inference. However, the model inference is probably maintained for service in a long time, while the collaboration, especially when the clients belong to different organizations, is unpredictable in real-world scenarios, such as concellation of contract, network unavailablity, etc., resulting in the failure of them. To address this issue, we, at the first attempt, propose a flexible Active-Passive Federated learning (APFed) framework. Specifically, the active client is the initiator of a learning task and responsible to build the complete model, while the passive clients only serve as assistants. Once the model built, the active client can make inference independently. In addition, we instance the APFed framework into two classification methods with employing the reconstruction loss and the contrastive loss on passive clients, respectively. Meanwhile, the two methods are tested in a set of experiments and achieves desired results, validating their effectiveness.
FlexCare: Leveraging Cross-Task Synergy for Flexible Multimodal Healthcare Prediction
Jun 17, 2024

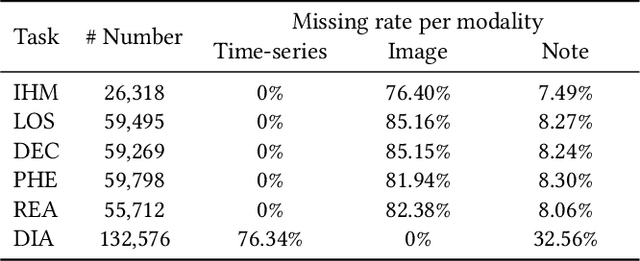
Abstract:Multimodal electronic health record (EHR) data can offer a holistic assessment of a patient's health status, supporting various predictive healthcare tasks. Recently, several studies have embraced the multitask learning approach in the healthcare domain, exploiting the inherent correlations among clinical tasks to predict multiple outcomes simultaneously. However, existing methods necessitate samples to possess complete labels for all tasks, which places heavy demands on the data and restricts the flexibility of the model. Meanwhile, within a multitask framework with multimodal inputs, how to comprehensively consider the information disparity among modalities and among tasks still remains a challenging problem. To tackle these issues, a unified healthcare prediction model, also named by \textbf{FlexCare}, is proposed to flexibly accommodate incomplete multimodal inputs, promoting the adaption to multiple healthcare tasks. The proposed model breaks the conventional paradigm of parallel multitask prediction by decomposing it into a series of asynchronous single-task prediction. Specifically, a task-agnostic multimodal information extraction module is presented to capture decorrelated representations of diverse intra- and inter-modality patterns. Taking full account of the information disparities between different modalities and different tasks, we present a task-guided hierarchical multimodal fusion module that integrates the refined modality-level representations into an individual patient-level representation. Experimental results on multiple tasks from MIMIC-IV/MIMIC-CXR/MIMIC-NOTE datasets demonstrate the effectiveness of the proposed method. Additionally, further analysis underscores the feasibility and potential of employing such a multitask strategy in the healthcare domain. The source code is available at https://github.com/mhxu1998/FlexCare.
UniCompress: Enhancing Multi-Data Medical Image Compression with Knowledge Distillation
May 27, 2024Abstract:In the field of medical image compression, Implicit Neural Representation (INR) networks have shown remarkable versatility due to their flexible compression ratios, yet they are constrained by a one-to-one fitting approach that results in lengthy encoding times. Our novel method, ``\textbf{UniCompress}'', innovatively extends the compression capabilities of INR by being the first to compress multiple medical data blocks using a single INR network. By employing wavelet transforms and quantization, we introduce a codebook containing frequency domain information as a prior input to the INR network. This enhances the representational power of INR and provides distinctive conditioning for different image blocks. Furthermore, our research introduces a new technique for the knowledge distillation of implicit representations, simplifying complex model knowledge into more manageable formats to improve compression ratios. Extensive testing on CT and electron microscopy (EM) datasets has demonstrated that UniCompress outperforms traditional INR methods and commercial compression solutions like HEVC, especially in complex and high compression scenarios. Notably, compared to existing INR techniques, UniCompress achieves a 4$\sim$5 times increase in compression speed, marking a significant advancement in the field of medical image compression. Codes will be publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge