Amina Asif
An Aggregation of Aggregation Methods in Computational Pathology
Nov 02, 2022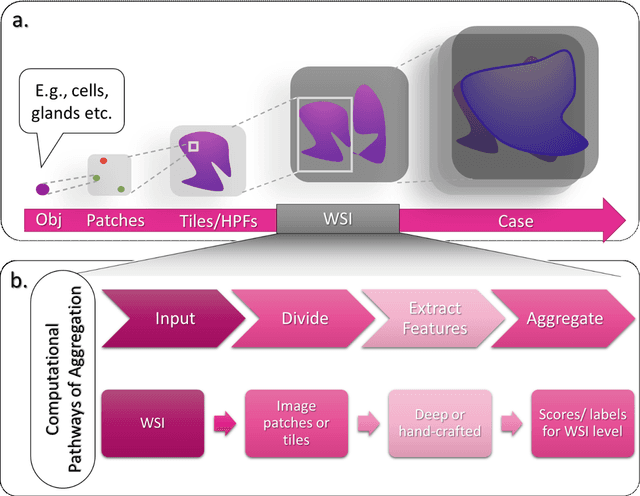
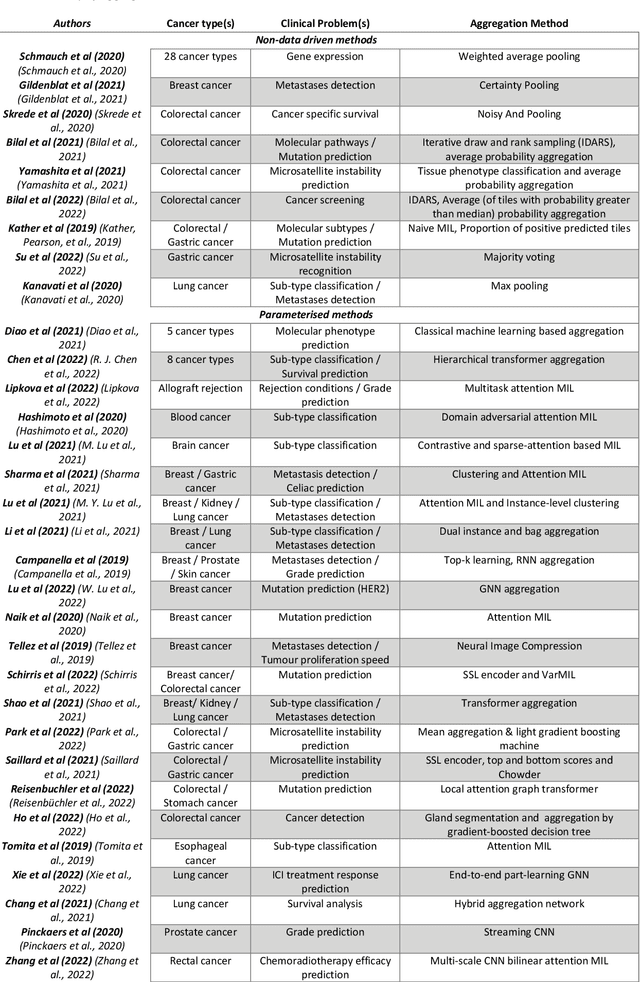
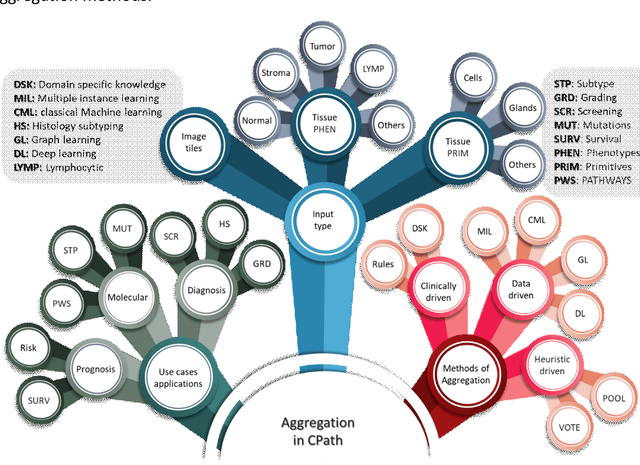
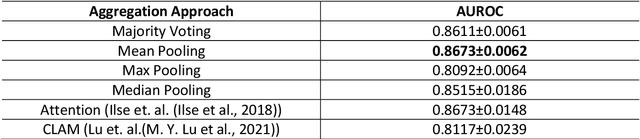
Abstract:Image analysis and machine learning algorithms operating on multi-gigapixel whole-slide images (WSIs) often process a large number of tiles (sub-images) and require aggregating predictions from the tiles in order to predict WSI-level labels. In this paper, we present a review of existing literature on various types of aggregation methods with a view to help guide future research in the area of computational pathology (CPath). We propose a general CPath workflow with three pathways that consider multiple levels and types of data and the nature of computation to analyse WSIs for predictive modelling. We categorize aggregation methods according to the context and representation of the data, features of computational modules and CPath use cases. We compare and contrast different methods based on the principle of multiple instance learning, perhaps the most commonly used aggregation method, covering a wide range of CPath literature. To provide a fair comparison, we consider a specific WSI-level prediction task and compare various aggregation methods for that task. Finally, we conclude with a list of objectives and desirable attributes of aggregation methods in general, pros and cons of the various approaches, some recommendations and possible future directions.
Rank the triplets: A ranking-based multiple instance learning framework for detecting HPV infection in head and neck cancers using routine H&E images
Jun 16, 2022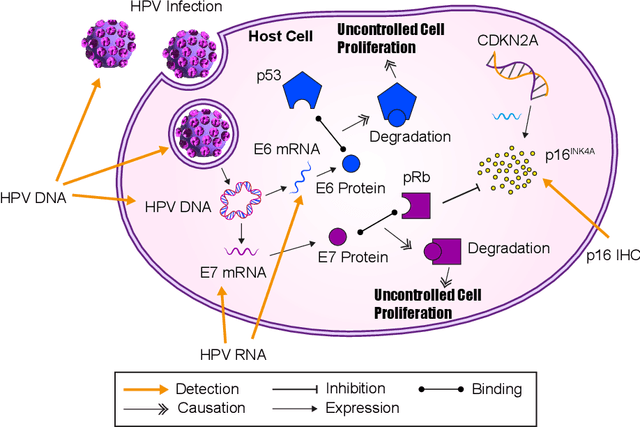

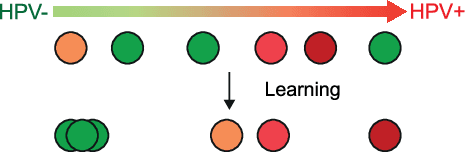
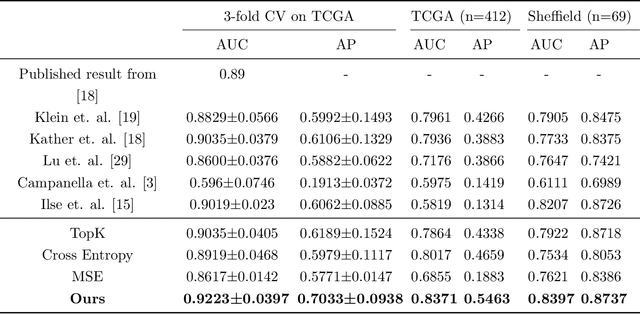
Abstract:The aetiology of head and neck squamous cell carcinoma (HNSCC) involves multiple carcinogens such as alcohol, tobacco and infection with human papillomavirus (HPV). As the HPV infection influences the prognosis, treatment and survival of patients with HNSCC, it is important to determine the HPV status of these tumours. In this paper, we propose a novel triplet-ranking loss function and a multiple instance learning pipeline for HPV status prediction. This achieves a new state-of-the-art performance in HPV detection using only the routine H&E stained WSIs on two HNSCC cohorts. Furthermore, a comprehensive tumour microenvironment profiling was performed, which characterised the unique patterns between HPV+/- HNSCC from genomic, immunology and cellular perspectives. Positive correlations of the proposed score with different subtypes of T cells (e.g. T cells follicular helper, CD8+ T cells), and negative correlations with macrophages and connective cells (e.g. fibroblast) were identified, which is in line with clinical findings. Unique gene expression profiles were also identified with respect to HPV infection status, and is in line with existing findings.
REET: Robustness Evaluation and Enhancement Toolbox for Computational Pathology
Jan 28, 2022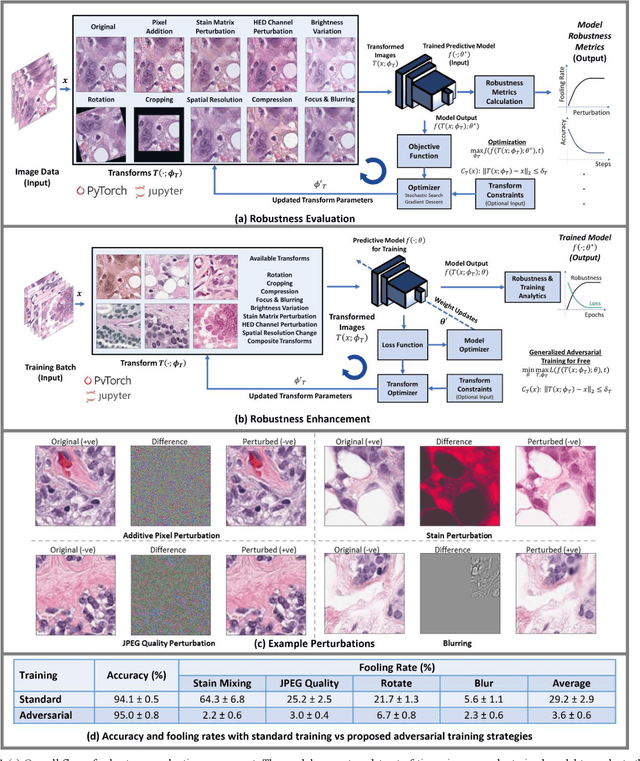
Abstract:Motivation: Digitization of pathology laboratories through digital slide scanners and advances in deep learning approaches for objective histological assessment have resulted in rapid progress in the field of computational pathology (CPath) with wide-ranging applications in medical and pharmaceutical research as well as clinical workflows. However, the estimation of robustness of CPath models to variations in input images is an open problem with a significant impact on the down-stream practical applicability, deployment and acceptability of these approaches. Furthermore, development of domain-specific strategies for enhancement of robustness of such models is of prime importance as well. Implementation and Availability: In this work, we propose the first domain-specific Robustness Evaluation and Enhancement Toolbox (REET) for computational pathology applications. It provides a suite of algorithmic strategies for enabling robustness assessment of predictive models with respect to specialized image transformations such as staining, compression, focusing, blurring, changes in spatial resolution, brightness variations, geometric changes as well as pixel-level adversarial perturbations. Furthermore, REET also enables efficient and robust training of deep learning pipelines in computational pathology. REET is implemented in Python and is available at the following URL: https://github.com/alexjfoote/reetoolbox. Contact: Fayyaz.minhas@warwick.ac.uk
Towards Launching AI Algorithms for Cellular Pathology into Clinical & Pharmaceutical Orbits
Dec 17, 2021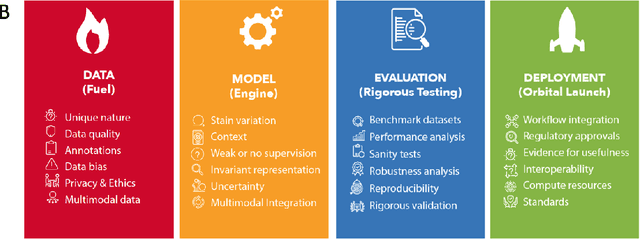

Abstract:Computational Pathology (CPath) is an emerging field concerned with the study of tissue pathology via computational algorithms for the processing and analysis of digitized high-resolution images of tissue slides. Recent deep learning based developments in CPath have successfully leveraged sheer volume of raw pixel data in histology images for predicting target parameters in the domains of diagnostics, prognostics, treatment sensitivity and patient stratification -- heralding the promise of a new data-driven AI era for both histopathology and oncology. With data serving as the fuel and AI as the engine, CPath algorithms are poised to be ready for takeoff and eventual launch into clinical and pharmaceutical orbits. In this paper, we discuss CPath limitations and associated challenges to enable the readers distinguish hope from hype and provide directions for future research to overcome some of the major challenges faced by this budding field to enable its launch into the two orbits.
Now You See It, Now You Dont: Adversarial Vulnerabilities in Computational Pathology
Jun 16, 2021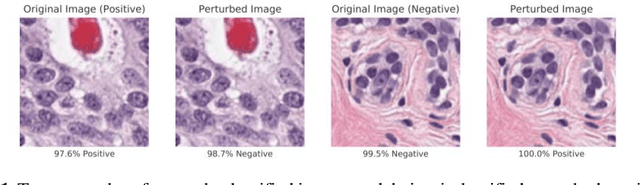

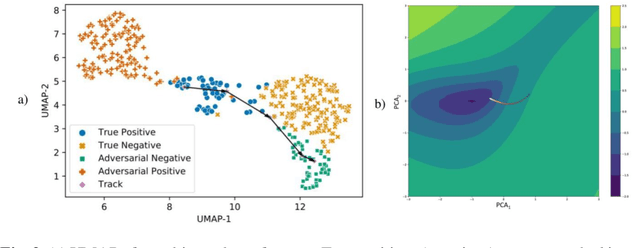
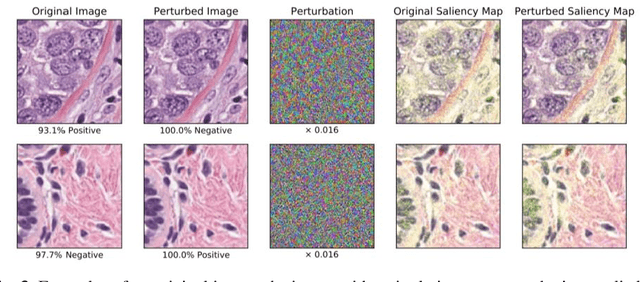
Abstract:Deep learning models are routinely employed in computational pathology (CPath) for solving problems of diagnostic and prognostic significance. Typically, the generalization performance of CPath models is analyzed using evaluation protocols such as cross-validation and testing on multi-centric cohorts. However, to ensure that such CPath solutions are robust and safe for use in a clinical setting, a critical analysis of their predictive performance and vulnerability to adversarial attacks is required, which is the focus of this paper. Specifically, we show that a highly accurate model for classification of tumour patches in pathology images (AUC > 0.95) can easily be attacked with minimal perturbations which are imperceptible to lay humans and trained pathologists alike. Our analytical results show that it is possible to generate single-instance white-box attacks on specific input images with high success rate and low perturbation energy. Furthermore, we have also generated a single universal perturbation matrix using the training dataset only which, when added to unseen test images, results in forcing the trained neural network to flip its prediction labels with high confidence at a success rate of > 84%. We systematically analyze the relationship between perturbation energy of an adversarial attack, its impact on morphological constructs of clinical significance, their perceptibility by a trained pathologist and saliency maps obtained using deep learning models. Based on our analysis, we strongly recommend that computational pathology models be critically analyzed using the proposed adversarial validation strategy prior to clinical adoption.
Learning Neural Activations
Dec 27, 2019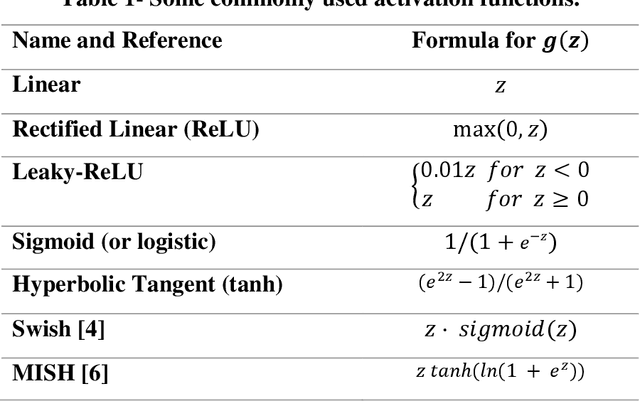
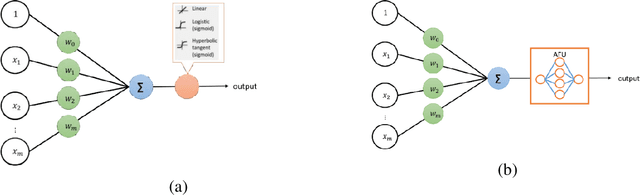
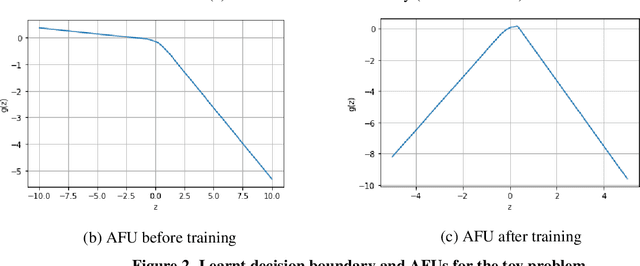
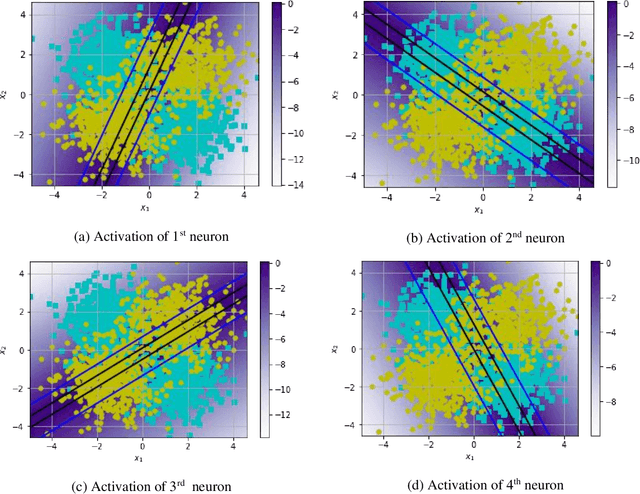
Abstract:An artificial neuron is modelled as a weighted summation followed by an activation function which determines its output. A wide variety of activation functions such as rectified linear units (ReLU), leaky-ReLU, Swish, MISH, etc. have been explored in the literature. In this short paper, we explore what happens when the activation function of each neuron in an artificial neural network is learned natively from data alone. This is achieved by modelling the activation function of each neuron as a small neural network whose weights are shared by all neurons in the original network. We list our primary findings in the conclusions section. The code for our analysis is available at: https://github.com/amina01/Learning-Neural-Activations.
Generalized Learning with Rejection for Classification and Regression Problems
Nov 03, 2019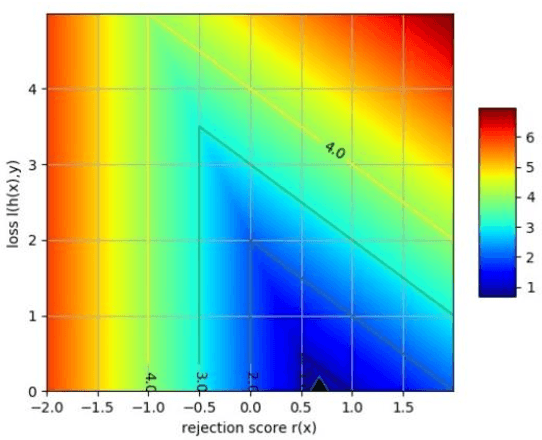

Abstract:Learning with rejection (LWR) allows development of machine learning systems with the ability to discard low confidence decisions generated by a prediction model. That is, just like human experts, LWR allows machine models to abstain from generating a prediction when reliability of the prediction is expected to be low. Several frameworks for this learning with rejection have been proposed in the literature. However, most of them work for classification problems only and regression with rejection has not been studied in much detail. In this work, we present a neural framework for LWR based on a generalized meta-loss function that involves simultaneous training of two neural network models: a predictor model for generating predictions and a rejecter model for deciding whether the prediction should be accepted or rejected. The proposed framework can be used for classification as well as regression and other related machine learning tasks. We have demonstrated the applicability and effectiveness of the method on synthetically generated data as well as benchmark datasets from UCI machine learning repository for both classification and regression problems. Despite being simpler in implementation, the proposed scheme for learning with rejection has shown to perform at par or better than previously proposed methods. Furthermore, we have applied the method to the problem of hurricane intensity prediction from satellite imagery. Significant improvement in performance as compared to conventional supervised methods shows the effectiveness of the proposed scheme in real-world regression problems.
An embarrassingly simple approach to neural multiple instance classification
May 06, 2019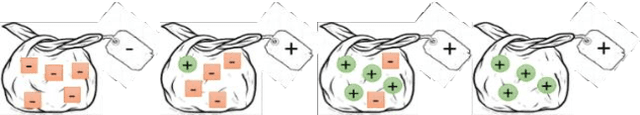
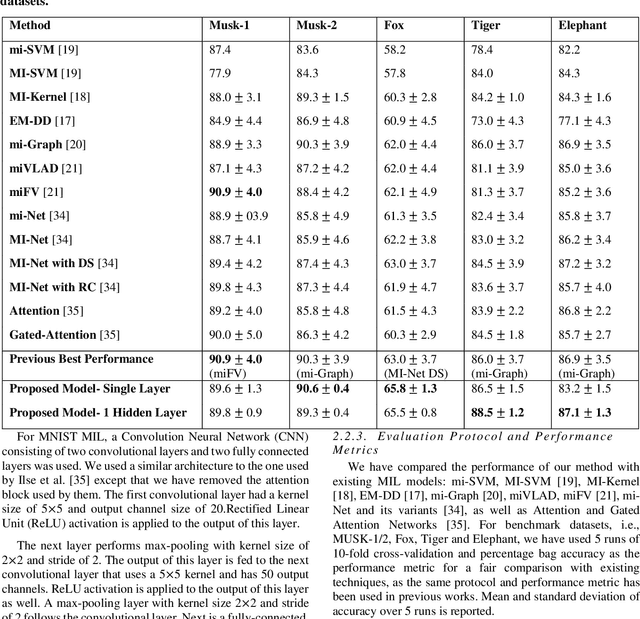
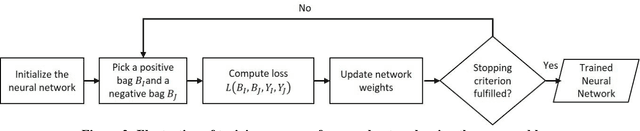
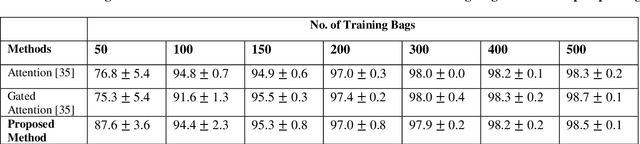
Abstract:Multiple Instance Learning (MIL) is a weak supervision learning paradigm that allows modeling of machine learning problems in which labels are available only for groups of examples called bags. A positive bag may contain one or more positive examples but it is not known which examples in the bag are positive. All examples in a negative bag belong to the negative class. Such problems arise frequently in fields of computer vision, medical image processing and bioinformatics. Many neural network based solutions have been proposed in the literature for MIL, however, almost all of them rely on introducing specialized blocks and connectivity in the architectures. In this paper, we present a novel and effective approach to Multiple Instance Learning in neural networks. Instead of making changes to the architectures, we propose a simple bag-level ranking loss function that allows Multiple Instance Classification in any neural architecture. We have demonstrated the effectiveness of our proposed method for popular MIL benchmark datasets. In addition, we have tested the performance of our method in convolutional neural networks used to model an MIL problem derived from the well-known MNIST dataset. Results have shown that despite being simpler, our proposed scheme is comparable or better than existing methods in the literature in practical scenarios. Python code files for all the experiments can be found at https://github.com/amina01/ESMIL.
* 7 pages
Ten ways to fool the masses with machine learning
Jan 07, 2019
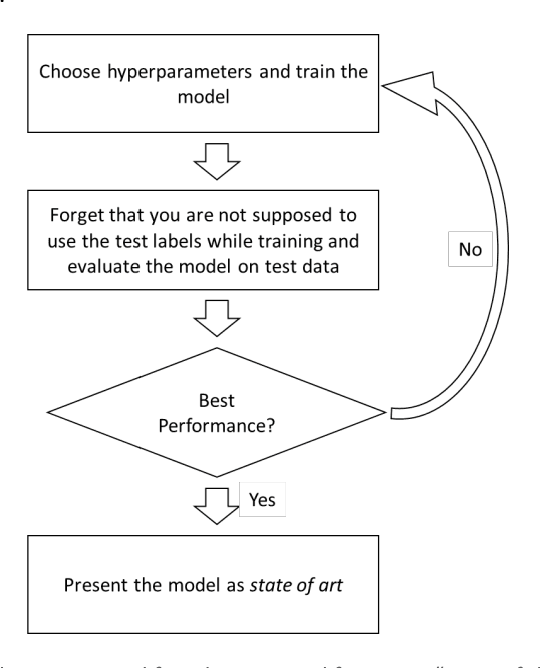
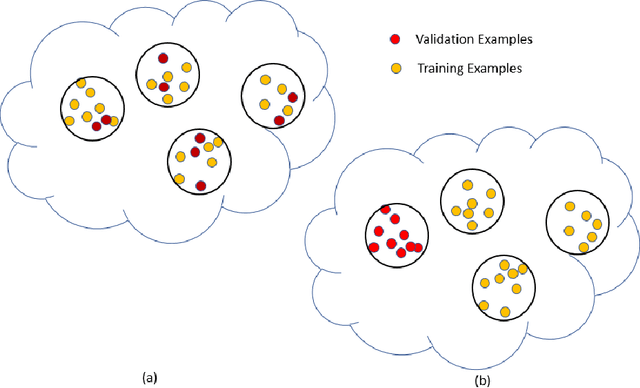
Abstract:If you want to tell people the truth, make them laugh, otherwise they'll kill you. (source unclear) Machine learning and deep learning are the technologies of the day for developing intelligent automatic systems. However, a key hurdle for progress in the field is the literature itself: we often encounter papers that report results that are difficult to reconstruct or reproduce, results that mis-represent the performance of the system, or contain other biases that limit their validity. In this semi-humorous article, we discuss issues that arise in running and reporting results of machine learning experiments. The purpose of the article is to provide a list of watch out points for researchers to be aware of when developing machine learning models or writing and reviewing machine learning papers.
A generalized meta-loss function for distillation and learning using privileged information for classification and regression
Nov 16, 2018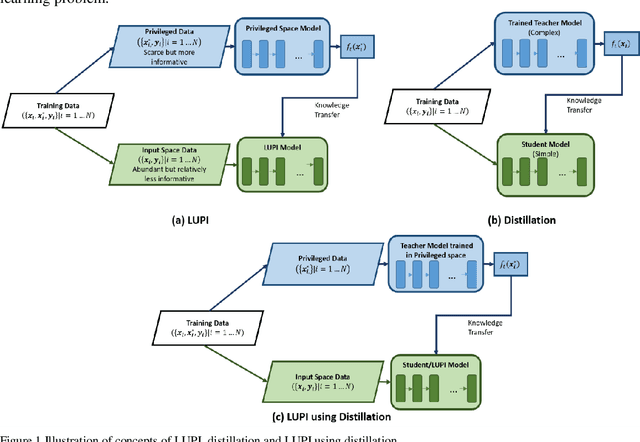
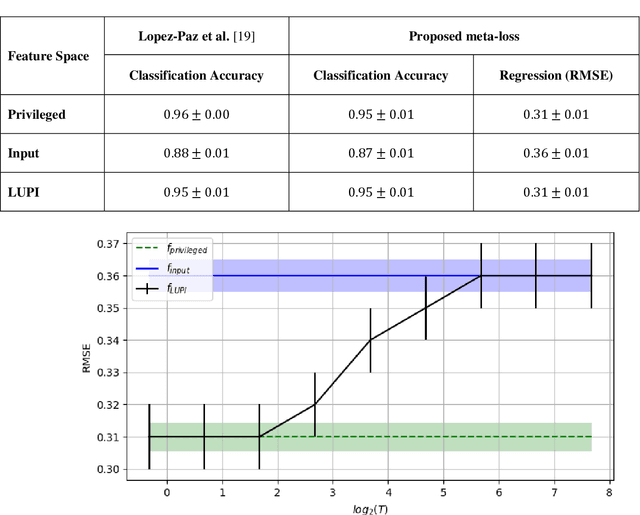
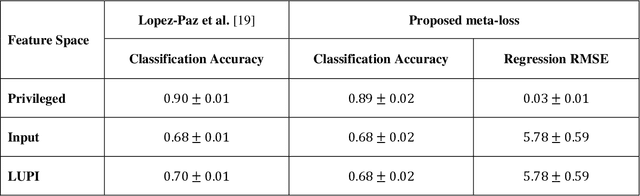
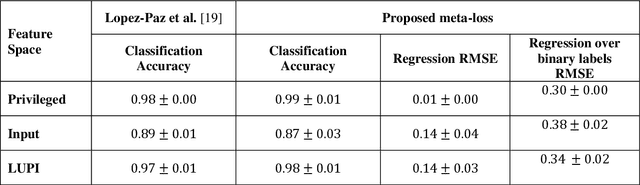
Abstract:Learning using privileged information and distillation are powerful machine learning frameworks that allow a machine learning model to be learned from an existing model or from a classifier trained over another feature space. Existing implementations of learning using privileged information are limited to classification only. In this work, we have proposed a novel meta-loss function that allows the general application of learning using privileged information and distillation to not only classification but also regression and other related problems. Our experimental results show the usefulness of the proposed scheme.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge