Alex Foote
Tackling Polysemanticity with Neuron Embeddings
Nov 12, 2024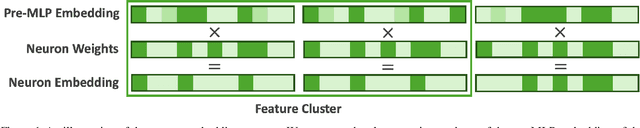


Abstract:We present neuron embeddings, a representation that can be used to tackle polysemanticity by identifying the distinct semantic behaviours in a neuron's characteristic dataset examples, making downstream manual or automatic interpretation much easier. We apply our method to GPT2-small, and provide a UI for exploring the results. Neuron embeddings are computed using a model's internal representations and weights, making them domain and architecture agnostic and removing the risk of introducing external structure which may not reflect a model's actual computation. We describe how neuron embeddings can be used to measure neuron polysemanticity, which could be applied to better evaluate the efficacy of Sparse Auto-Encoders (SAEs).
Neuron to Graph: Interpreting Language Model Neurons at Scale
May 31, 2023



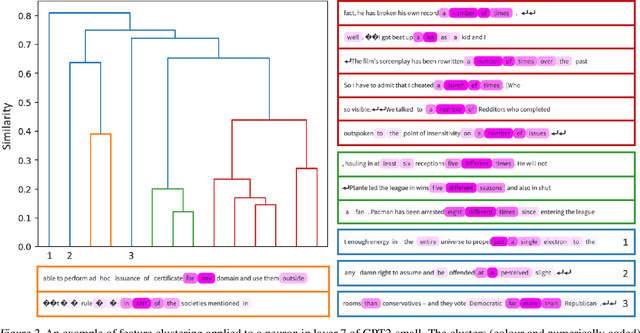
Abstract:Advances in Large Language Models (LLMs) have led to remarkable capabilities, yet their inner mechanisms remain largely unknown. To understand these models, we need to unravel the functions of individual neurons and their contribution to the network. This paper introduces a novel automated approach designed to scale interpretability techniques across a vast array of neurons within LLMs, to make them more interpretable and ultimately safe. Conventional methods require examination of examples with strong neuron activation and manual identification of patterns to decipher the concepts a neuron responds to. We propose Neuron to Graph (N2G), an innovative tool that automatically extracts a neuron's behaviour from the dataset it was trained on and translates it into an interpretable graph. N2G uses truncation and saliency methods to emphasise only the most pertinent tokens to a neuron while enriching dataset examples with diverse samples to better encompass the full spectrum of neuron behaviour. These graphs can be visualised to aid researchers' manual interpretation, and can generate token activations on text for automatic validation by comparison with the neuron's ground truth activations, which we use to show that the model is better at predicting neuron activation than two baseline methods. We also demonstrate how the generated graph representations can be flexibly used to facilitate further automation of interpretability research, by searching for neurons with particular properties, or programmatically comparing neurons to each other to identify similar neurons. Our method easily scales to build graph representations for all neurons in a 6-layer Transformer model using a single Tesla T4 GPU, allowing for wide usability. We release the code and instructions for use at https://github.com/alexjfoote/Neuron2Graph.
N2G: A Scalable Approach for Quantifying Interpretable Neuron Representations in Large Language Models
Apr 22, 2023



Abstract:Understanding the function of individual neurons within language models is essential for mechanistic interpretability research. We propose $\textbf{Neuron to Graph (N2G)}$, a tool which takes a neuron and its dataset examples, and automatically distills the neuron's behaviour on those examples to an interpretable graph. This presents a less labour intensive approach to interpreting neurons than current manual methods, that will better scale these methods to Large Language Models (LLMs). We use truncation and saliency methods to only present the important tokens, and augment the dataset examples with more diverse samples to better capture the extent of neuron behaviour. These graphs can be visualised to aid manual interpretation by researchers, but can also output token activations on text to compare to the neuron's ground truth activations for automatic validation. N2G represents a step towards scalable interpretability methods by allowing us to convert neurons in an LLM to interpretable representations of measurable quality.
REET: Robustness Evaluation and Enhancement Toolbox for Computational Pathology
Jan 28, 2022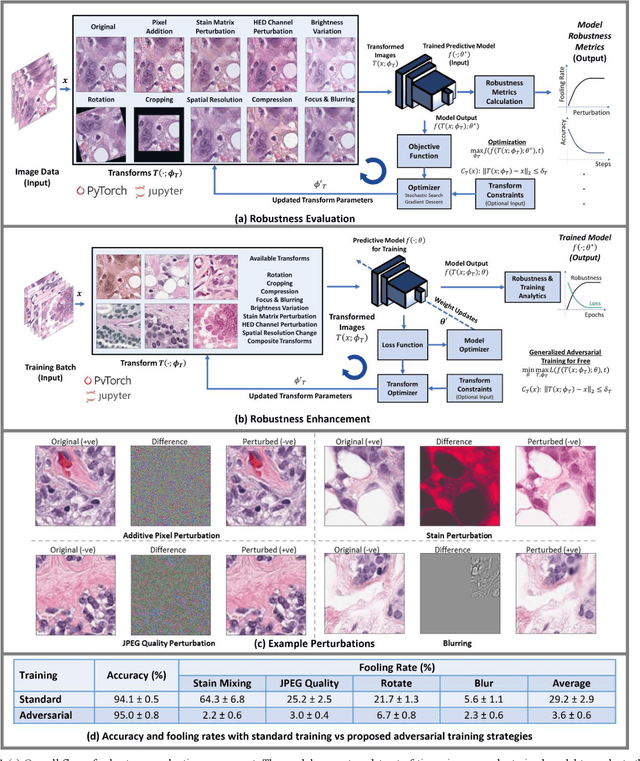
Abstract:Motivation: Digitization of pathology laboratories through digital slide scanners and advances in deep learning approaches for objective histological assessment have resulted in rapid progress in the field of computational pathology (CPath) with wide-ranging applications in medical and pharmaceutical research as well as clinical workflows. However, the estimation of robustness of CPath models to variations in input images is an open problem with a significant impact on the down-stream practical applicability, deployment and acceptability of these approaches. Furthermore, development of domain-specific strategies for enhancement of robustness of such models is of prime importance as well. Implementation and Availability: In this work, we propose the first domain-specific Robustness Evaluation and Enhancement Toolbox (REET) for computational pathology applications. It provides a suite of algorithmic strategies for enabling robustness assessment of predictive models with respect to specialized image transformations such as staining, compression, focusing, blurring, changes in spatial resolution, brightness variations, geometric changes as well as pixel-level adversarial perturbations. Furthermore, REET also enables efficient and robust training of deep learning pipelines in computational pathology. REET is implemented in Python and is available at the following URL: https://github.com/alexjfoote/reetoolbox. Contact: Fayyaz.minhas@warwick.ac.uk
Now You See It, Now You Dont: Adversarial Vulnerabilities in Computational Pathology
Jun 16, 2021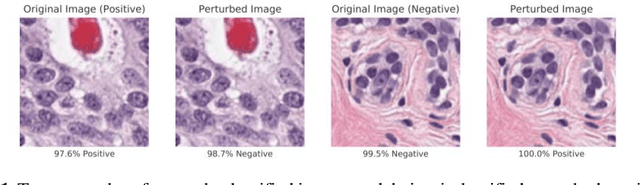

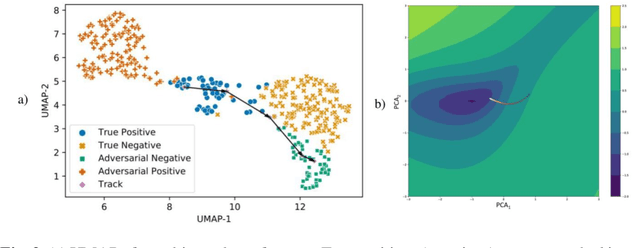
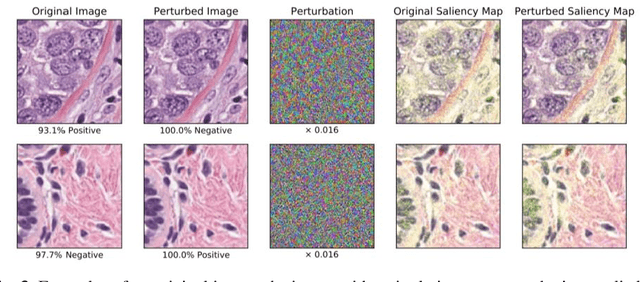
Abstract:Deep learning models are routinely employed in computational pathology (CPath) for solving problems of diagnostic and prognostic significance. Typically, the generalization performance of CPath models is analyzed using evaluation protocols such as cross-validation and testing on multi-centric cohorts. However, to ensure that such CPath solutions are robust and safe for use in a clinical setting, a critical analysis of their predictive performance and vulnerability to adversarial attacks is required, which is the focus of this paper. Specifically, we show that a highly accurate model for classification of tumour patches in pathology images (AUC > 0.95) can easily be attacked with minimal perturbations which are imperceptible to lay humans and trained pathologists alike. Our analytical results show that it is possible to generate single-instance white-box attacks on specific input images with high success rate and low perturbation energy. Furthermore, we have also generated a single universal perturbation matrix using the training dataset only which, when added to unseen test images, results in forcing the trained neural network to flip its prediction labels with high confidence at a success rate of > 84%. We systematically analyze the relationship between perturbation energy of an adversarial attack, its impact on morphological constructs of clinical significance, their perceptibility by a trained pathologist and saliency maps obtained using deep learning models. Based on our analysis, we strongly recommend that computational pathology models be critically analyzed using the proposed adversarial validation strategy prior to clinical adoption.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge