Asa Ben-Hur
Department of Computer Science, Colorado State University, Fort Collins, USA
Insights into performance evaluation of com-pound-protein interaction prediction methods
Jan 28, 2022
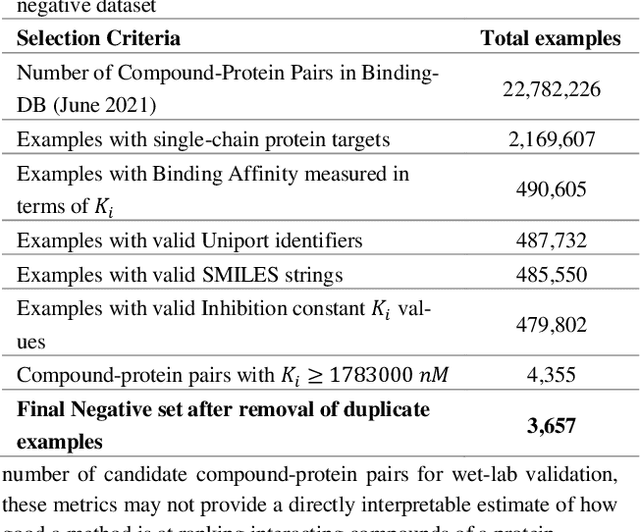
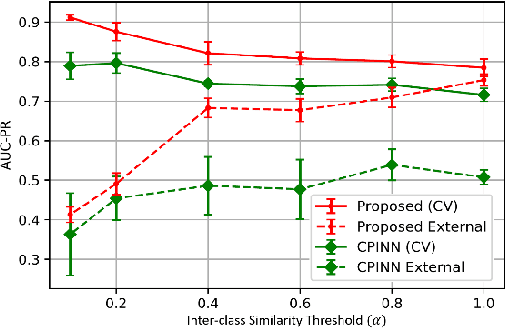
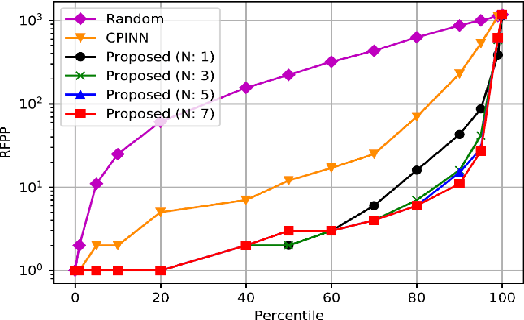
Abstract:Motivation: Machine learning based prediction of compound-protein interactions (CPIs) is important for drug design, screening and repurposing studies and can improve the efficiency and cost-effectiveness of wet lab assays. Despite the publication of many research papers reporting CPI predictors in the recent years, we have observed a number of fundamental issues in experiment design that lead to over optimistic estimates of model performance. Results: In this paper, we analyze the impact of several important factors affecting generalization perfor-mance of CPI predictors that are overlooked in existing work: 1. Similarity between training and test examples in cross-validation 2. The strategy for generating negative examples, in the absence of experimentally verified negative examples. 3. Choice of evaluation protocols and performance metrics and their alignment with real-world use of CPI predictors in screening large compound libraries. Using both an existing state-of-the-art method (CPI-NN) and a proposed kernel based approach, we have found that assessment of predictive performance of CPI predictors requires careful con-trol over similarity between training and test examples. We also show that random pairing for gen-erating synthetic negative examples for training and performance evaluation results in models with better generalization performance in comparison to more sophisticated strategies used in existing studies. Furthermore, we have found that our kernel based approach, despite its simple design, exceeds the prediction performance of CPI-NN. We have used the proposed model for compound screening of several proteins including SARS-CoV-2 Spike and Human ACE2 proteins and found strong evidence in support of its top hits. Availability: Code and raw experimental results available at https://github.com/adibayaseen/HKRCPI Contact: Fayyaz.minhas@warwick.ac.uk
Ten ways to fool the masses with machine learning
Jan 07, 2019
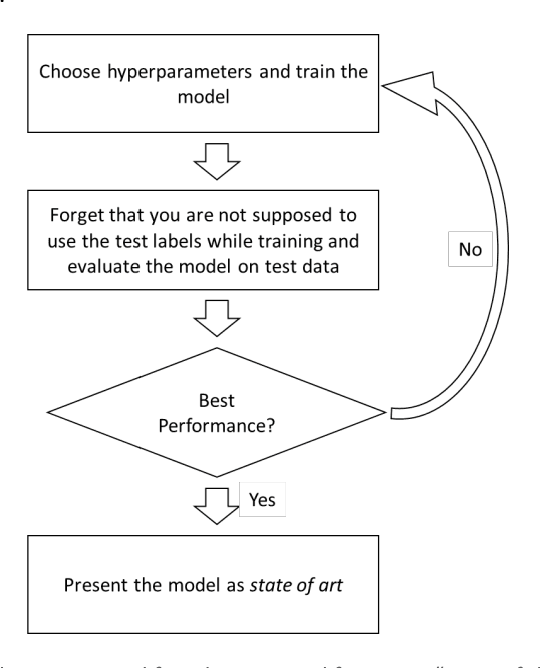
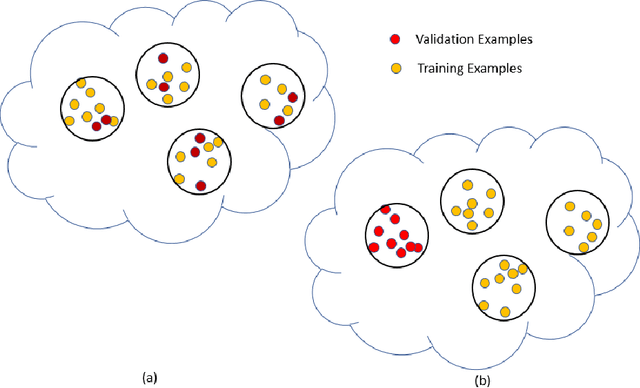
Abstract:If you want to tell people the truth, make them laugh, otherwise they'll kill you. (source unclear) Machine learning and deep learning are the technologies of the day for developing intelligent automatic systems. However, a key hurdle for progress in the field is the literature itself: we often encounter papers that report results that are difficult to reconstruct or reproduce, results that mis-represent the performance of the system, or contain other biases that limit their validity. In this semi-humorous article, we discuss issues that arise in running and reporting results of machine learning experiments. The purpose of the article is to provide a list of watch out points for researchers to be aware of when developing machine learning models or writing and reviewing machine learning papers.
pyLEMMINGS: Large Margin Multiple Instance Classification and Ranking for Bioinformatics Applications
Nov 14, 2017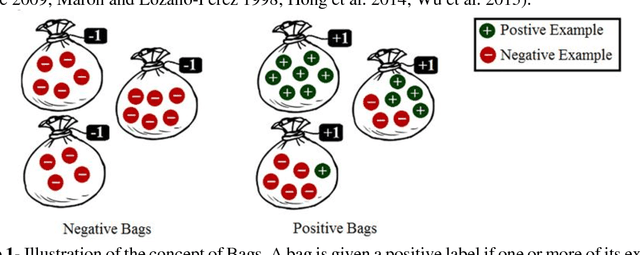

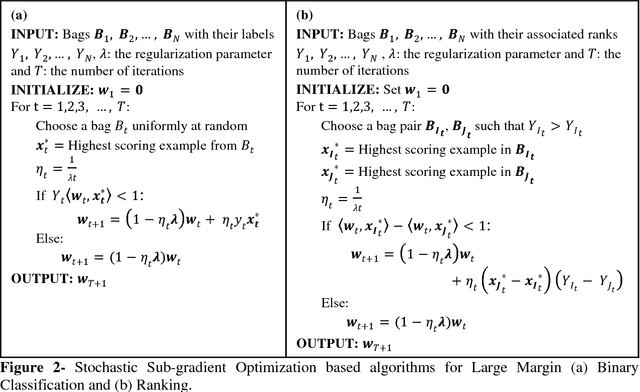
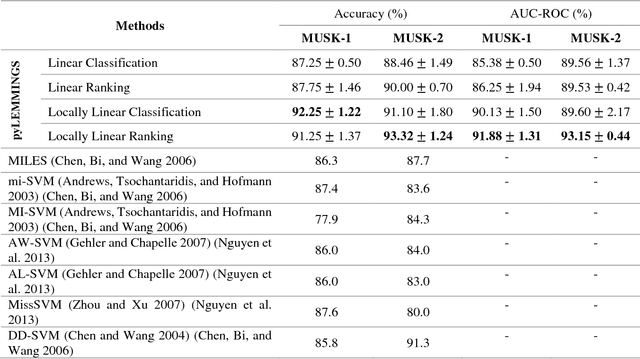
Abstract:Motivation: A major challenge in the development of machine learning based methods in computational biology is that data may not be accurately labeled due to the time and resources required for experimentally annotating properties of proteins and DNA sequences. Standard supervised learning algorithms assume accurate instance-level labeling of training data. Multiple instance learning is a paradigm for handling such labeling ambiguities. However, the widely used large-margin classification methods for multiple instance learning are heuristic in nature with high computational requirements. In this paper, we present stochastic sub-gradient optimization large margin algorithms for multiple instance classification and ranking, and provide them in a software suite called pyLEMMINGS. Results: We have tested pyLEMMINGS on a number of bioinformatics problems as well as benchmark datasets. pyLEMMINGS has successfully been able to identify functionally important segments of proteins: binding sites in Calmodulin binding proteins, prion forming regions, and amyloid cores. pyLEMMINGS achieves state-of-the-art performance in all these tasks, demonstrating the value of multiple instance learning. Furthermore, our method has shown more than 100-fold improvement in terms of running time as compared to heuristic solutions with improved accuracy over benchmark datasets. Availability and Implementation: pyLEMMINGS python package is available for download at: http://faculty.pieas.edu.pk/fayyaz/software.html#pylemmings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge