Fayyaz ul Amir Afsar Minhas
DepoRanker: A Web Tool to predict Klebsiella Depolymerases using Machine Learning
Jan 27, 2025Abstract:Background: Phage therapy shows promise for treating antibiotic-resistant Klebsiella infections. Identifying phage depolymerases that target Klebsiella capsular polysaccharides is crucial, as these capsules contribute to biofilm formation and virulence. However, homology-based searches have limitations in novel depolymerase discovery. Objective: To develop a machine learning model for identifying and ranking potential phage depolymerases targeting Klebsiella. Methods: We developed DepoRanker, a machine learning algorithm to rank proteins by their likelihood of being depolymerases. The model was experimentally validated on 5 newly characterized proteins and compared to BLAST. Results: DepoRanker demonstrated superior performance to BLAST in identifying potential depolymerases. Experimental validation confirmed its predictive ability on novel proteins. Conclusions: DepoRanker provides an accurate and functional tool to expedite depolymerase discovery for phage therapy against Klebsiella. It is available as a webserver and open-source software. Availability: Webserver: https://deporanker.dcs.warwick.ac.uk/ Source code: https://github.com/wgrgwrght/deporanker
HistoKernel: Whole Slide Image Level Maximum Mean Discrepancy Kernels for Pan-Cancer Predictive Modelling
Aug 09, 2024



Abstract:Machine learning in computational pathology (CPath) often aggregates patch-level predictions from multi-gigapixel Whole Slide Images (WSIs) to generate WSI-level prediction scores for crucial tasks such as survival prediction and drug effect prediction. However, current methods do not explicitly characterize distributional differences between patch sets within WSIs. We introduce HistoKernel, a novel Maximum Mean Discrepancy (MMD) kernel that measures distributional similarity between WSIs for enhanced prediction performance on downstream prediction tasks. Our comprehensive analysis demonstrates HistoKernel's effectiveness across various machine learning tasks, including retrieval (n = 9,362), drug sensitivity regression (n = 551), point mutation classification (n = 3,419), and survival analysis (n = 2,291), outperforming existing deep learning methods. Additionally, HistoKernel seamlessly integrates multi-modal data and offers a novel perturbation-based method for patch-level explainability. This work pioneers the use of kernel-based methods for WSI-level predictive modeling, opening new avenues for research. Code is available at https://github.com/pkeller00/HistoKernel.
Maximum Mean Discrepancy Kernels for Predictive and Prognostic Modeling of Whole Slide Images
Jan 23, 2023Abstract:How similar are two images? In computational pathology, where Whole Slide Images (WSIs) of digitally scanned tissue samples from patients can be multi-gigapixels in size, determination of degree of similarity between two WSIs is a challenging task with a number of practical applications. In this work, we explore a novel strategy based on kernelized Maximum Mean Discrepancy (MMD) analysis for determination of pairwise similarity between WSIs. The proposed approach works by calculating MMD between two WSIs using kernels over deep features of image patches. This allows representation of an entire dataset of WSIs as a kernel matrix for WSI level clustering, weakly-supervised prediction of TP-53 mutation status in breast cancer patients from their routine WSIs as well as survival analysis with state of the art prediction performance. We believe that this work will open up further avenues for application of WSI-level kernels for predictive and prognostic tasks in computational pathology.
ALBRT: Cellular Composition Prediction in Routine Histology Images
Aug 26, 2021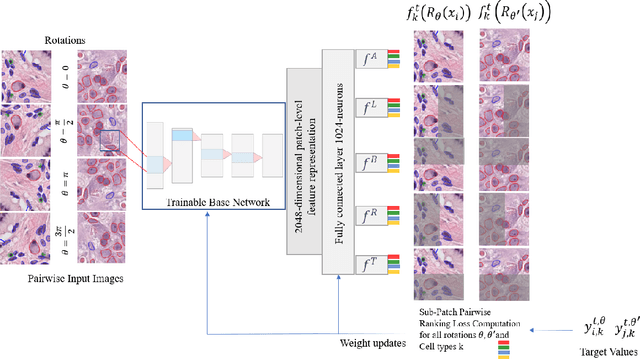
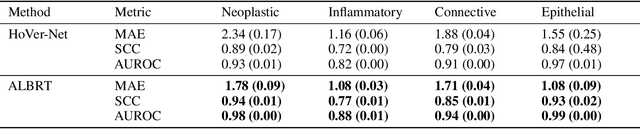
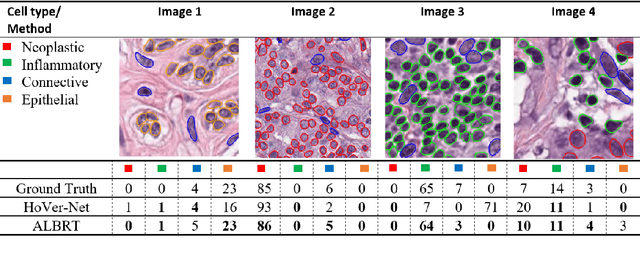
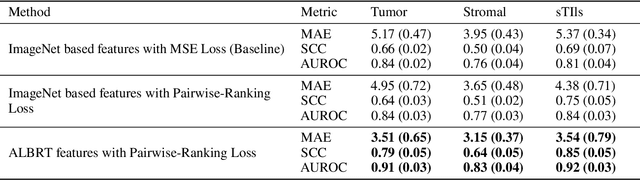
Abstract:Cellular composition prediction, i.e., predicting the presence and counts of different types of cells in the tumor microenvironment from a digitized image of a Hematoxylin and Eosin (H&E) stained tissue section can be used for various tasks in computational pathology such as the analysis of cellular topology and interactions, subtype prediction, survival analysis, etc. In this work, we propose an image-based cellular composition predictor (ALBRT) which can accurately predict the presence and counts of different types of cells in a given image patch. ALBRT, by its contrastive-learning inspired design, learns a compact and rotation-invariant feature representation that is then used for cellular composition prediction of different cell types. It offers significant improvement over existing state-of-the-art approaches for cell classification and counting. The patch-level feature representation learned by ALBRT is transferrable for cellular composition analysis over novel datasets and can also be utilized for downstream prediction tasks in CPath as well. The code and the inference webserver for the proposed method are available at the URL: https://github.com/engrodawood/ALBRT.
All You Need is Color: Image based Spatial Gene Expression Prediction using Neural Stain Learning
Aug 26, 2021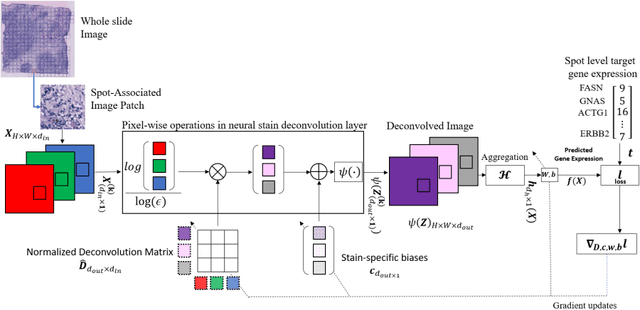
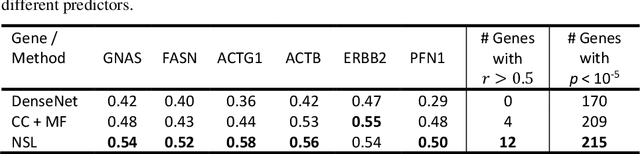
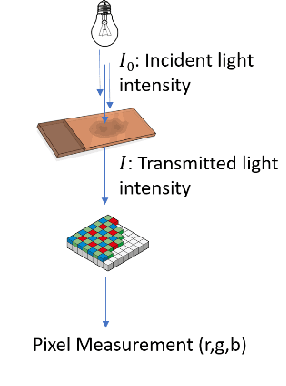
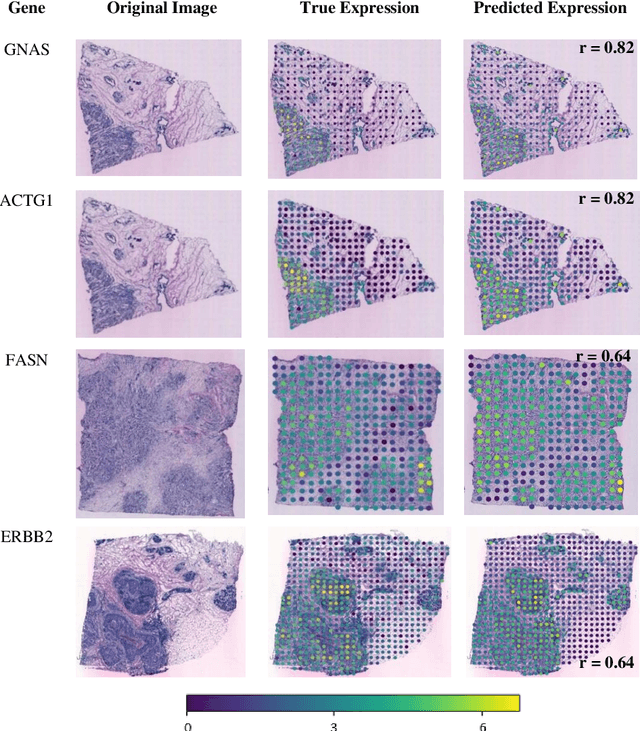
Abstract:"Is it possible to predict expression levels of different genes at a given spatial location in the routine histology image of a tumor section by modeling its stain absorption characteristics?" In this work, we propose a "stain-aware" machine learning approach for prediction of spatial transcriptomic gene expression profiles using digital pathology image of a routine Hematoxylin & Eosin (H&E) histology section. Unlike recent deep learning methods which are used for gene expression prediction, our proposed approach termed Neural Stain Learning (NSL) explicitly models the association of stain absorption characteristics of the tissue with gene expression patterns in spatial transcriptomics by learning a problem-specific stain deconvolution matrix in an end-to-end manner. The proposed method with only 11 trainable weight parameters outperforms both classical regression models with cellular composition and morphological features as well as deep learning methods. We have found that the gene expression predictions from the proposed approach show higher correlations with true expression values obtained through sequencing for a larger set of genes in comparison to other approaches.
Learning Neural Activations
Dec 27, 2019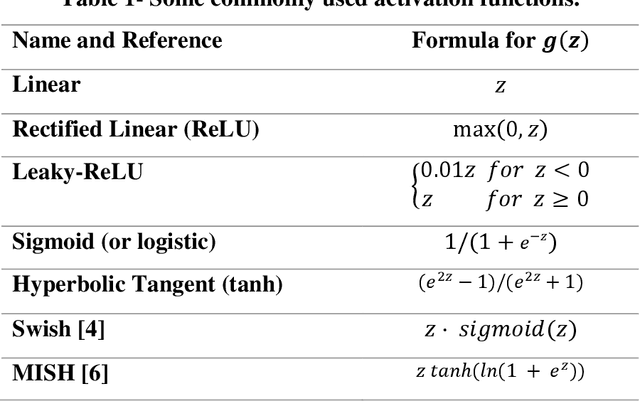
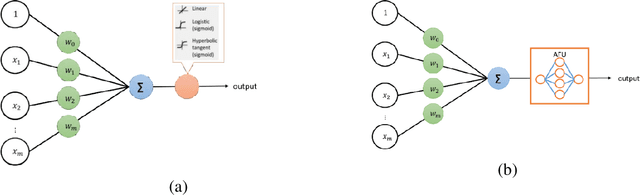
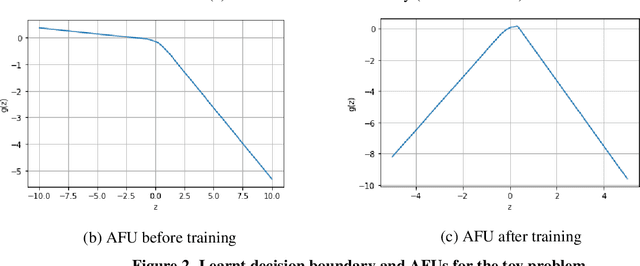
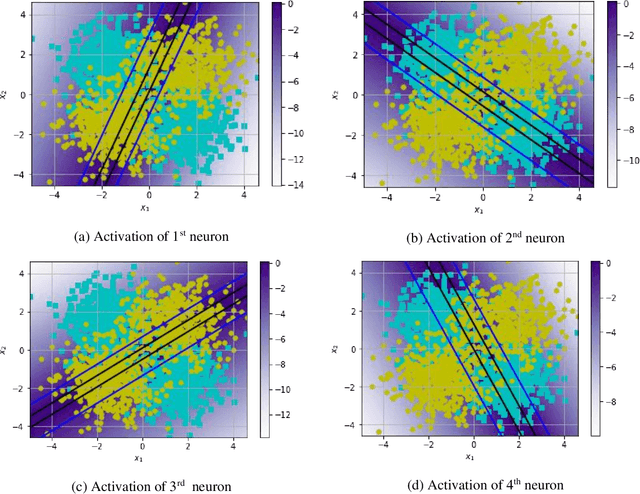
Abstract:An artificial neuron is modelled as a weighted summation followed by an activation function which determines its output. A wide variety of activation functions such as rectified linear units (ReLU), leaky-ReLU, Swish, MISH, etc. have been explored in the literature. In this short paper, we explore what happens when the activation function of each neuron in an artificial neural network is learned natively from data alone. This is achieved by modelling the activation function of each neuron as a small neural network whose weights are shared by all neurons in the original network. We list our primary findings in the conclusions section. The code for our analysis is available at: https://github.com/amina01/Learning-Neural-Activations.
Generalized Learning with Rejection for Classification and Regression Problems
Nov 03, 2019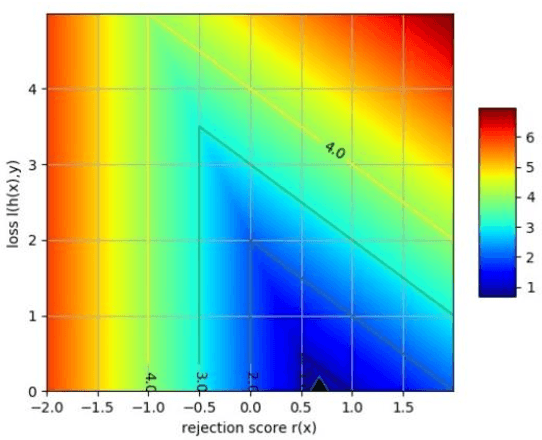

Abstract:Learning with rejection (LWR) allows development of machine learning systems with the ability to discard low confidence decisions generated by a prediction model. That is, just like human experts, LWR allows machine models to abstain from generating a prediction when reliability of the prediction is expected to be low. Several frameworks for this learning with rejection have been proposed in the literature. However, most of them work for classification problems only and regression with rejection has not been studied in much detail. In this work, we present a neural framework for LWR based on a generalized meta-loss function that involves simultaneous training of two neural network models: a predictor model for generating predictions and a rejecter model for deciding whether the prediction should be accepted or rejected. The proposed framework can be used for classification as well as regression and other related machine learning tasks. We have demonstrated the applicability and effectiveness of the method on synthetically generated data as well as benchmark datasets from UCI machine learning repository for both classification and regression problems. Despite being simpler in implementation, the proposed scheme for learning with rejection has shown to perform at par or better than previously proposed methods. Furthermore, we have applied the method to the problem of hurricane intensity prediction from satellite imagery. Significant improvement in performance as compared to conventional supervised methods shows the effectiveness of the proposed scheme in real-world regression problems.
An embarrassingly simple approach to neural multiple instance classification
May 06, 2019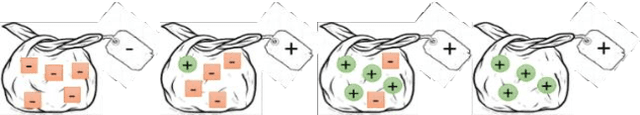
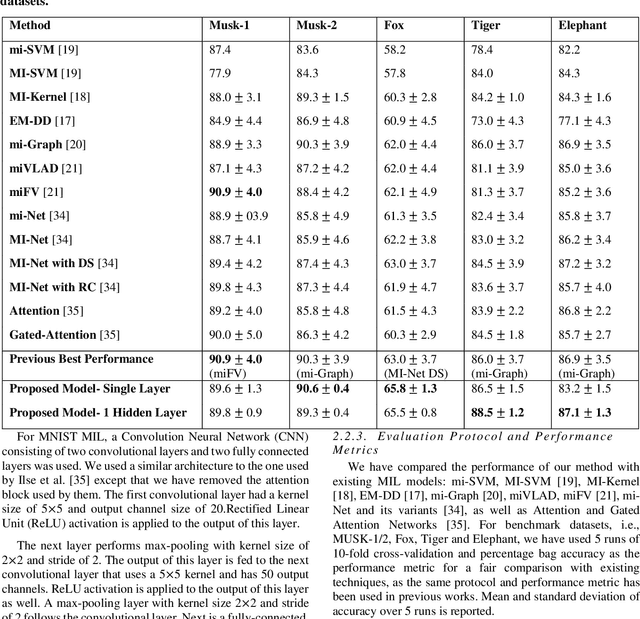
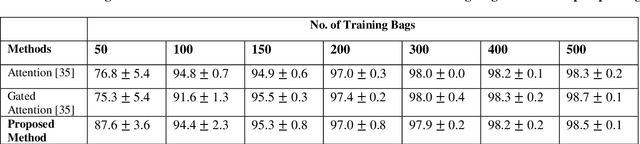
Abstract:Multiple Instance Learning (MIL) is a weak supervision learning paradigm that allows modeling of machine learning problems in which labels are available only for groups of examples called bags. A positive bag may contain one or more positive examples but it is not known which examples in the bag are positive. All examples in a negative bag belong to the negative class. Such problems arise frequently in fields of computer vision, medical image processing and bioinformatics. Many neural network based solutions have been proposed in the literature for MIL, however, almost all of them rely on introducing specialized blocks and connectivity in the architectures. In this paper, we present a novel and effective approach to Multiple Instance Learning in neural networks. Instead of making changes to the architectures, we propose a simple bag-level ranking loss function that allows Multiple Instance Classification in any neural architecture. We have demonstrated the effectiveness of our proposed method for popular MIL benchmark datasets. In addition, we have tested the performance of our method in convolutional neural networks used to model an MIL problem derived from the well-known MNIST dataset. Results have shown that despite being simpler, our proposed scheme is comparable or better than existing methods in the literature in practical scenarios. Python code files for all the experiments can be found at https://github.com/amina01/ESMIL.
* 7 pages
A generalized meta-loss function for distillation and learning using privileged information for classification and regression
Nov 16, 2018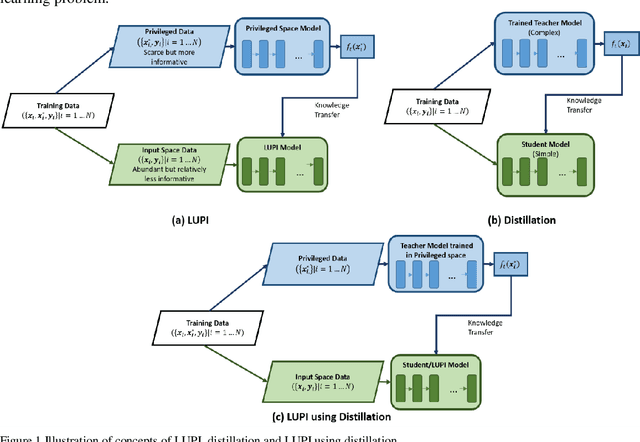
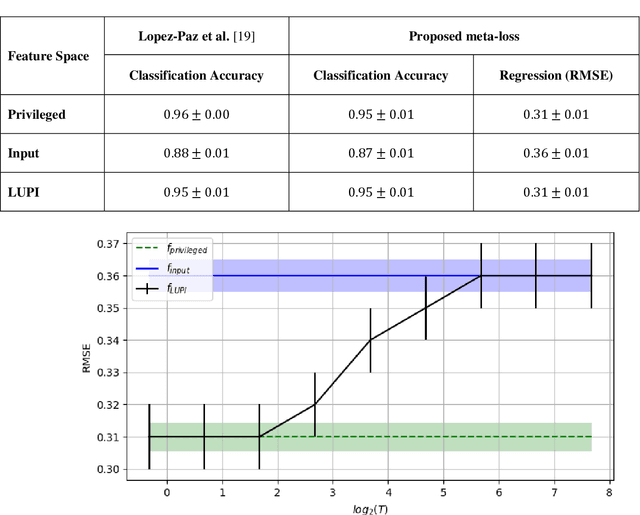
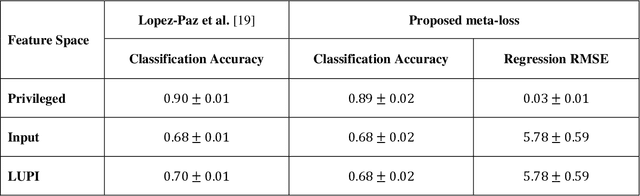
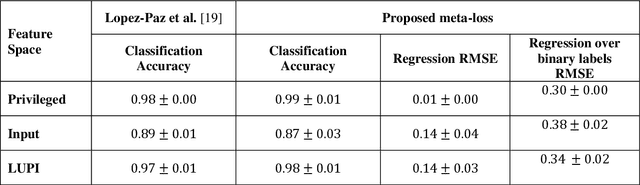
Abstract:Learning using privileged information and distillation are powerful machine learning frameworks that allow a machine learning model to be learned from an existing model or from a classifier trained over another feature space. Existing implementations of learning using privileged information are limited to classification only. In this work, we have proposed a novel meta-loss function that allows the general application of learning using privileged information and distillation to not only classification but also regression and other related problems. Our experimental results show the usefulness of the proposed scheme.
pyLEMMINGS: Large Margin Multiple Instance Classification and Ranking for Bioinformatics Applications
Nov 14, 2017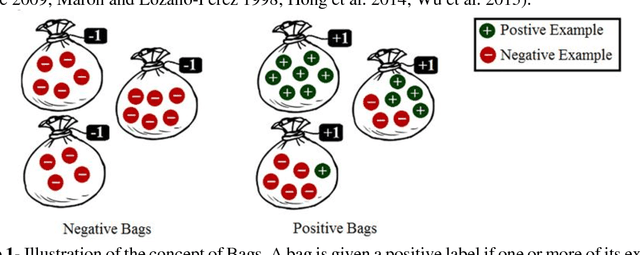

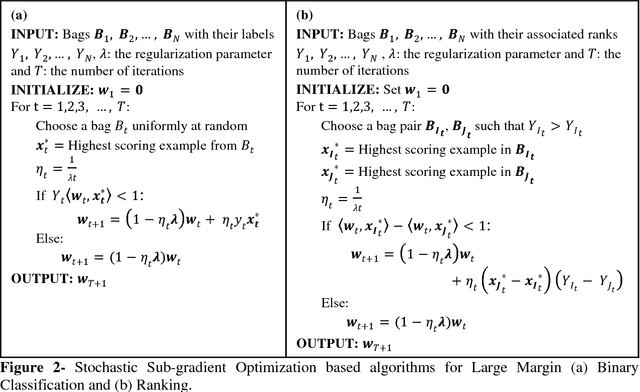
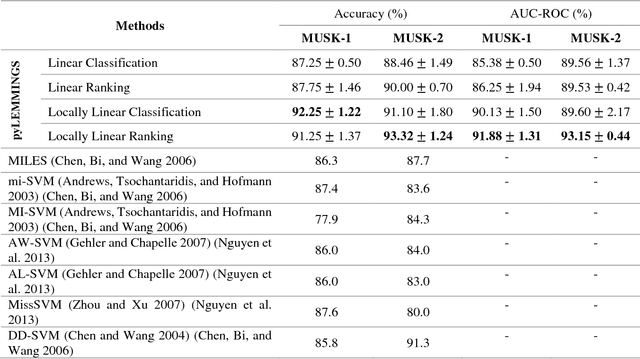
Abstract:Motivation: A major challenge in the development of machine learning based methods in computational biology is that data may not be accurately labeled due to the time and resources required for experimentally annotating properties of proteins and DNA sequences. Standard supervised learning algorithms assume accurate instance-level labeling of training data. Multiple instance learning is a paradigm for handling such labeling ambiguities. However, the widely used large-margin classification methods for multiple instance learning are heuristic in nature with high computational requirements. In this paper, we present stochastic sub-gradient optimization large margin algorithms for multiple instance classification and ranking, and provide them in a software suite called pyLEMMINGS. Results: We have tested pyLEMMINGS on a number of bioinformatics problems as well as benchmark datasets. pyLEMMINGS has successfully been able to identify functionally important segments of proteins: binding sites in Calmodulin binding proteins, prion forming regions, and amyloid cores. pyLEMMINGS achieves state-of-the-art performance in all these tasks, demonstrating the value of multiple instance learning. Furthermore, our method has shown more than 100-fold improvement in terms of running time as compared to heuristic solutions with improved accuracy over benchmark datasets. Availability and Implementation: pyLEMMINGS python package is available for download at: http://faculty.pieas.edu.pk/fayyaz/software.html#pylemmings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge