pyLEMMINGS: Large Margin Multiple Instance Classification and Ranking for Bioinformatics Applications
Paper and Code
Nov 14, 2017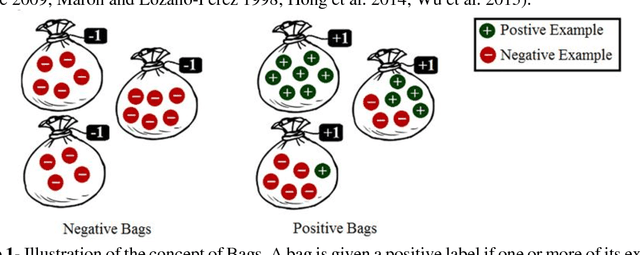

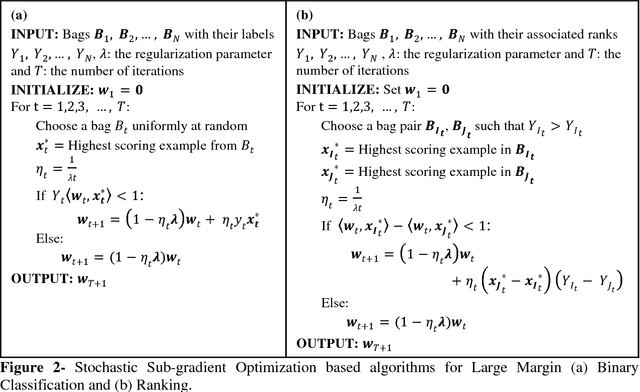
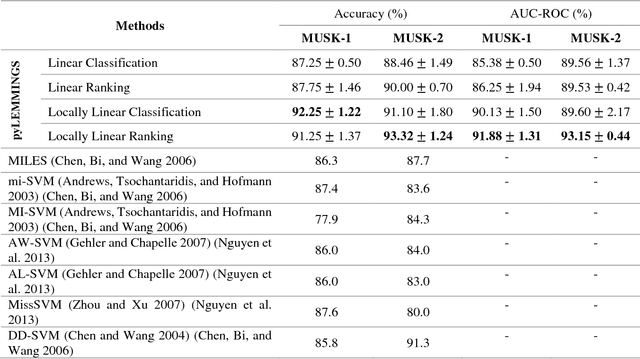
Motivation: A major challenge in the development of machine learning based methods in computational biology is that data may not be accurately labeled due to the time and resources required for experimentally annotating properties of proteins and DNA sequences. Standard supervised learning algorithms assume accurate instance-level labeling of training data. Multiple instance learning is a paradigm for handling such labeling ambiguities. However, the widely used large-margin classification methods for multiple instance learning are heuristic in nature with high computational requirements. In this paper, we present stochastic sub-gradient optimization large margin algorithms for multiple instance classification and ranking, and provide them in a software suite called pyLEMMINGS. Results: We have tested pyLEMMINGS on a number of bioinformatics problems as well as benchmark datasets. pyLEMMINGS has successfully been able to identify functionally important segments of proteins: binding sites in Calmodulin binding proteins, prion forming regions, and amyloid cores. pyLEMMINGS achieves state-of-the-art performance in all these tasks, demonstrating the value of multiple instance learning. Furthermore, our method has shown more than 100-fold improvement in terms of running time as compared to heuristic solutions with improved accuracy over benchmark datasets. Availability and Implementation: pyLEMMINGS python package is available for download at: http://faculty.pieas.edu.pk/fayyaz/software.html#pylemmings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge